The Association of HOXD-AS2 and MIR3142HG Gene Polymorphisms with Cervical Intraepithelial Neoplasia
-
摘要:
目的 探究靶向MAPK信号通路的HOXD-AS2、MIR3142HG基因多态性与宫颈上皮内瘤变(cervical intraepithelial neoplasia,CIN)的相关性。 方法 通过生物信息学工具筛选了2个单核苷酸多态性(single nucleotide polymorphism,SNP)位点,分别是位于HOXD-AS2启动子区域的rs1348808及位于MIR3142HG增强子区域的rs2431099。同时纳入了976名健康对照者和419名CIN患者,使用TaqMan探针法对其进行基因分型并分析与CIN的相关性。 结果 rs1348808的C等位基因可能是CIN3以及CIN2进展为CIN3的保护因素(OR = 0.77,95%CI:0.63~0.94;OR = 0.57,95%CI:0.37~0.90);rs2431099的A等位基因可能是CIN2的保护因素(OR = 0.50,95%CI:0.31~0.80)。 结论 HOXD-AS2、MIR3142HG的基因多态性可能与CIN相关。 Abstract:Objective To explore the association of HOXD-AS2 and MIR3142HG gene polymorphism with cervical intraepithelial neoplasia (CIN). Methods Two single nucleotide polymorphisms (SNPs) located in HOXD-AS2 (rs1348808) and located in MIR3142HG (rs2431099) were selected using bioinformatics tools. The two candidate SNPs were genotyped in 976 healthy individuals and 419 patients with CIN using the TaqMan probe method for genotyping, and the association with CIN were analyzed. Results rs1348808 C allele might be the protective factor for CIN3 as well as the progression from CIN2 to CIN3 (OR = 0.77, 95%CI: 0.63~0.94; OR = 0.57, 95%CI: 0.37~0.90). Similarly, rs2431099 A allele might be the protective factor of CIN2 (OR = 0.50, 95%CI: 0.31~0.80). Conclusion HOXD-AS2 and MIR3142HG gene polymorphisms might be associated with CIN. -
Key words:
- Cervical intraepithelial neoplasia /
- lncRNA /
- SNP /
- MAPK signaling pathway
-
鼻咽癌(nasopharyngeal carcinoma,NPC)来源于鼻咽部粘膜上皮,是我国南方高发肿瘤之一,因其位置隐匿,难以早期发现,故多数患者就诊时已处于III、IV期。鼻咽癌的治疗以放射治疗为主,联合化疗和/或分子靶向治疗,经综合治疗,仍有30%~40%的患者出现复发或远处转移,一旦出现复发和转移,其5 a生存率仅有25%到35%[1],化疗耐药和放疗抵抗是治疗失败的主要原因之一。对放、化疗后复发和转移的患者,再次行放、化疗效果较差,靶向治疗和免疫治疗价格昂贵、副作用较大、缓解时间较短。中药白藜芦醇(Resveratrol)在自然界中分布广泛、易于提取,它可以抑制包括鼻咽癌在内的多种肿瘤的增殖、促进肿瘤细胞凋亡,并可增加鼻咽癌对放射治疗和化学治疗的敏感性;可抑制口腔癌、乳腺癌、大肠癌、肺癌等多种肿瘤的转移[2-6],白藜芦醇是否能抑制鼻咽癌的转移,其可能的机制是什么,目前尚未见研究报导。本研究拟观察白藜芦醇对鼻咽癌转移能力的影响并初步探讨其影响鼻咽癌转移的分子机制,为鼻咽癌的治疗提供一种安全有效的治疗办法。
1. 材料与方法
1.1 实验材料
鼻咽癌细胞株CNE2来源于云南省肿瘤医院肿瘤研究所。Transwell迁移和侵袭小室购自美国Corning 有限公司。BCA法蛋白定量试剂盒购自北京普利莱基因技术有限公司。ECL化学发光检测试剂购自Millipore公司。CCK8试剂盒购自日本DOJINDO公司。MMP2、MMP9、p-Hsp27(ser82)抗体购自Cell Signaling technology,Inc。GAPDH、E-cadherin、Vimentin、Hsp27、Ezrin、p-Ezrin(T567)、Keratin18(角蛋白18)抗体均购自Abcam公司。白藜芦醇、二甲基亚砜(DMSO)、抗荧光淬灭封片剂(含DAPI)购自美国Sigma公司。
1.2 细胞增殖抑制(CCK-8)实验
取对数生长期鼻咽癌CNE2细胞,用胰酶消化为细胞悬液,接种于96孔板对应的孔内,每孔加入3000个细胞,然后将96孔板在培养箱中培养过夜。在96孔板中加入不同浓度的白藜芦醇,在培养箱内分别孵育24 h、36 h及48 h,每组设置5个副孔;用移液器吸去培养板中废液,然后加入100 μL无血清培养基到对应孔中,再向每孔加入10 μL CCK-8反应液;加完反应液后,将培养板放回培养箱内继续孵育2 h,使用酶标仪测定其在450 nm处的吸光度(OD值)。计算细胞增殖率:[(给药组OD值-对照组OD值)/对照组OD值]%。
1.3 Transwell小室试验
迁移实验:取对数生长期鼻咽癌CNE2细胞,用胰酶消化为细胞悬液,用细胞计数板将密度调整为5×105个/mL。向对应的孔中加入500 μL含15%FBS的1640,细胞悬液200 μL加入到Transwell迁移小室,然后将小室轻轻放入含15%FBS的1640培养基的对应孔中,放回孵箱,常规培养24 h;24 h后,弃去小室内液体,用棉签擦去小室内面的细胞,用PBS漂洗3次,置于4%多聚甲醛中固定20 min;再置于0.1%结晶紫中染色20 min;随后取下小室的底膜,将其贴在载玻片上,盖上盖玻片;每张膜随机取5个视野(×200),在显微镜下计数小室下室面的细胞数。侵袭实验:选用铺胶的侵袭小室,其余步骤同迁移实验。
1.4 免疫荧光实验
制备鼻咽癌CNE2细胞爬片,各细胞组经白藜芦醇处理后,按免疫荧光化学试验步骤处理,在湿盒中避光室温孵育2 h,用含抗荧光淬灭剂和DAPI的封片液封片,在荧光显微镜下观察并采集图像,观察NF-κB/p65的核转位情况。
1.5 WB实验
用不同浓度(0、10、20、40、80 μmol/L)的白藜芦醇处理鼻咽癌细胞CNE2,24 h后用RIPA裂解液提取蛋白,用BCA法定量蛋白浓度。取25 µg蛋白上样,经过SDS-PAGE凝胶电泳,转移至PVDF膜上,用5%的脱脂牛奶封闭液封闭2 h,加入一抗(1∶1000),在4 ℃下孵育过夜。TBST漂洗3次后,加入二抗(1∶2000)孵育1 h,TBST再次清洗后,使用AI600图像分析系统,拍照采集图像,采集图像后用Image Pro Plus软件计算各条带的灰度值,并计算目的基因与管家基因的比值,进行定量分析统计。
1.6 动物实验
选择SPF 级 BALB/C雌性裸鼠(4~5周左右,体重约18~22 g),严格饲养于SPF动物房层流架SPF环境中,恒温、恒湿、隔音、空气层流过滤,分笼饲养,每笼5只,灭菌水和饲料均由动物自由饮食用。动物饲养盒、空气过滤器、垫料、饲料和饮水等均经高压蒸汽灭菌,并定期更换。
裸鼠经适应性喂养1周后,取对数生长期鼻咽癌CNE2细胞,胰酶消化成细胞悬液,用细胞计数板计数后离心,每次均为1000 r/min,5 min,用PBS缓冲液洗涤3次,用 PBS缓冲液配制成2×105个细胞/只/30 μL的细胞悬液,置于冰盒中,1 h内按无菌操作要求用微量注射器注射入裸鼠左侧后肢的足掌皮下,每只裸鼠接种30 μL细胞悬液。将20只裸鼠根据白藜芦醇的给药浓度随机分为4组,即为0 mg/kg、25 mg/kg、50 mg/kg和100 mg/kg组,每组5只裸鼠。种瘤后第2天开始,治疗组每天给予不同浓度的白藜芦醇-羧甲基纤维素混悬液灌胃,给药量为小鼠体重(g)×20 μL/g,对照组根据小鼠体重给予相应体积的溶剂(0.5%羧甲基纤维素)。每天观察裸鼠活动、饮食等情况,每隔3 d测量裸鼠体重。自注射部位出现肉眼可见的肿瘤组织,开始记录最大长径(L)和横径(W),估算肿瘤体积(V),V = L×W2/2。至仁慈终点,将裸鼠麻醉,颈椎脱臼法处死,取材。取双侧腘窝淋巴结和足掌部肿瘤组织,拍照,测量大小和重量,置于4%多聚甲醛中固定,送至病理科石蜡包埋、切片后进行HE染色和免疫组化染色,显微镜下观察癌组织和Keratin18染色情况。
1.7 统计学处理
实验所得数据均采用SPSS22.0软件进行统计分析,实验至少重复3次。计量资料数据用均数±标准差表示,多个样本的均数比较用单因素方差分析,2组间的比较用t检验,P < 0.05为差异有统计学意义。
2. 结果
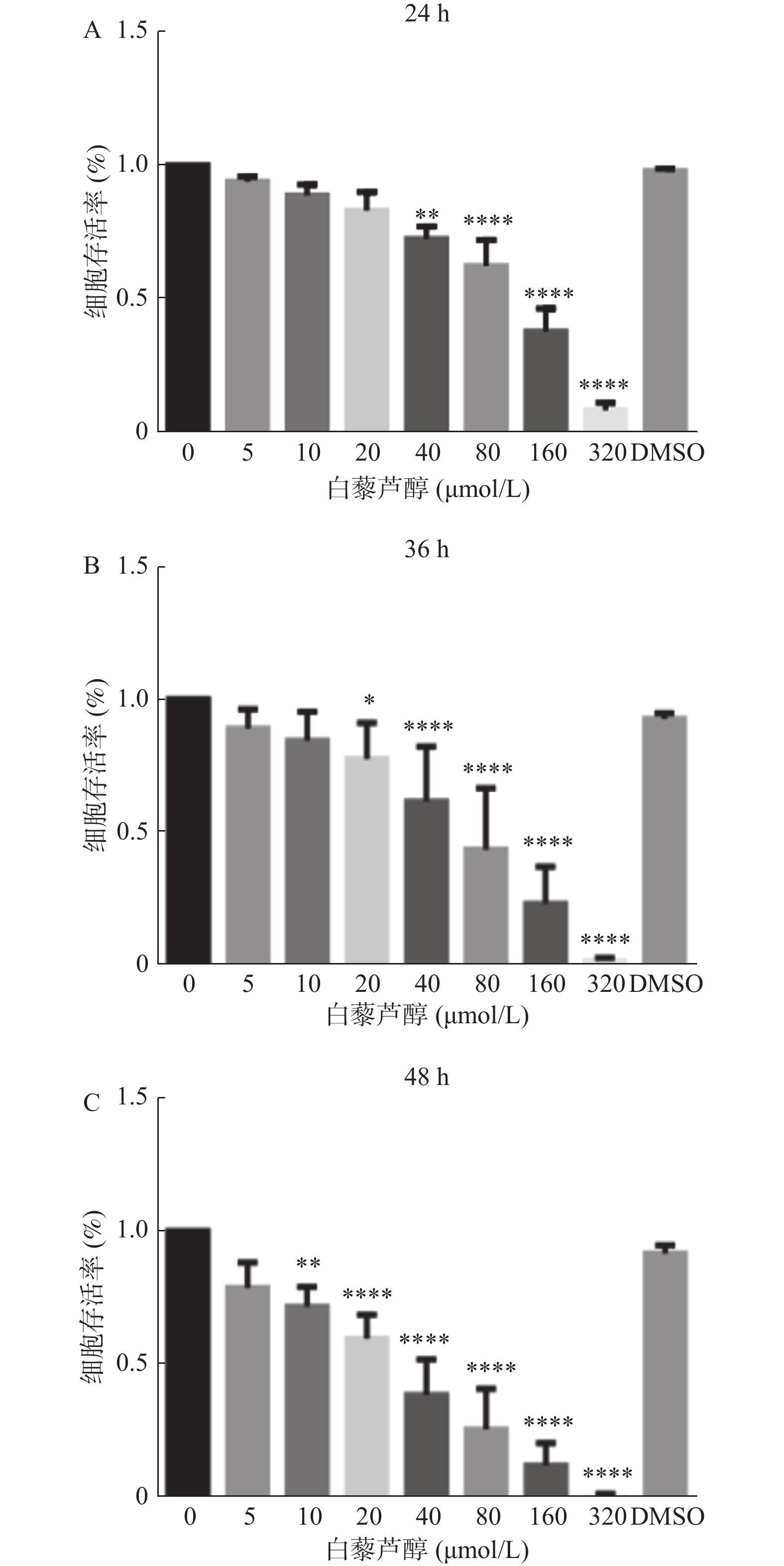
2.1 白藜芦醇可抑制鼻咽癌CNE2细胞的增殖能力
CCK8实验发现,白藜芦醇对鼻咽癌CNE2细胞的增殖有抑制作用,且抑制作用呈浓度和时间依赖性。当白藜芦醇浓度为80 μmol/L以下,作用于CNE2细胞24 h,白藜芦醇对CNE2细胞的抑制率未超过50%,图1。考虑到抑制率超过50%时存活细胞数量较少,可能影响到后续迁移和侵袭实验的结果,故选择0、10、20、40、80 μmol/L 5个不同浓度白藜芦醇处理CNE2细胞24 h以进行后续实验。
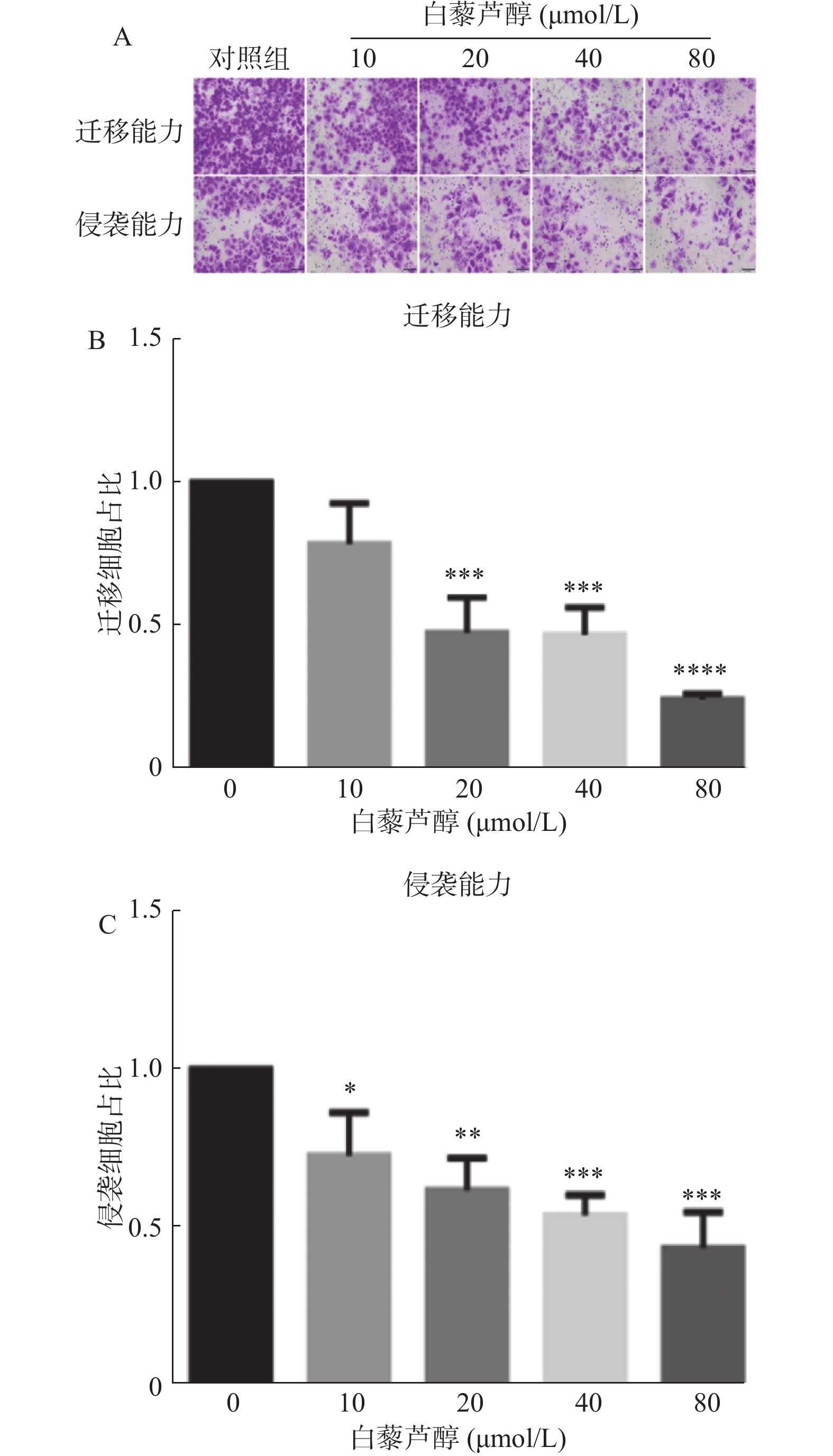
2.2 白藜芦醇明显抑制鼻咽癌CNE2细胞的侵袭、迁移能力
Transwell迁移侵袭实验发现,用10、20、40、80 μmol/L白藜芦醇处理CNE2细胞24 h后,各浓度组的穿膜细胞明显少于对照组(0 μmol/L组)见图2,随着白藜芦醇浓度的增高,对鼻咽癌CNE2细胞迁移和侵袭能力的抑制作用增强。
2.3 白藜芦醇可抑制鼻咽癌CNE2细胞中NF-κB/p65的核转位
CNE2细胞经过免疫荧光染色,红色代表NF-κB/p65,细胞核显示为蓝色。实验结果显示,白藜芦醇可能抑制NF-κB/p65的核转位,抑制作用呈浓度依赖性,见图3。
2.4 白藜芦醇可降低转移相关蛋白表达水平
Western blot检测发现,不同浓度(0、10、20、40、80 μmol/L)白藜芦醇处理鼻咽癌CNE2细胞24 h后:细胞内上皮表型标志物E-cadherin的表达水平上升(图4A);细胞内MMP2和MMP9的表达水平下调(图4B);细胞内磷酸化Ezrin(p-Ezrin)的表达水平明显下调(图4C);细胞内磷酸化Hsp27(p-Hsp27)的表达水平明显下调(图4D)。
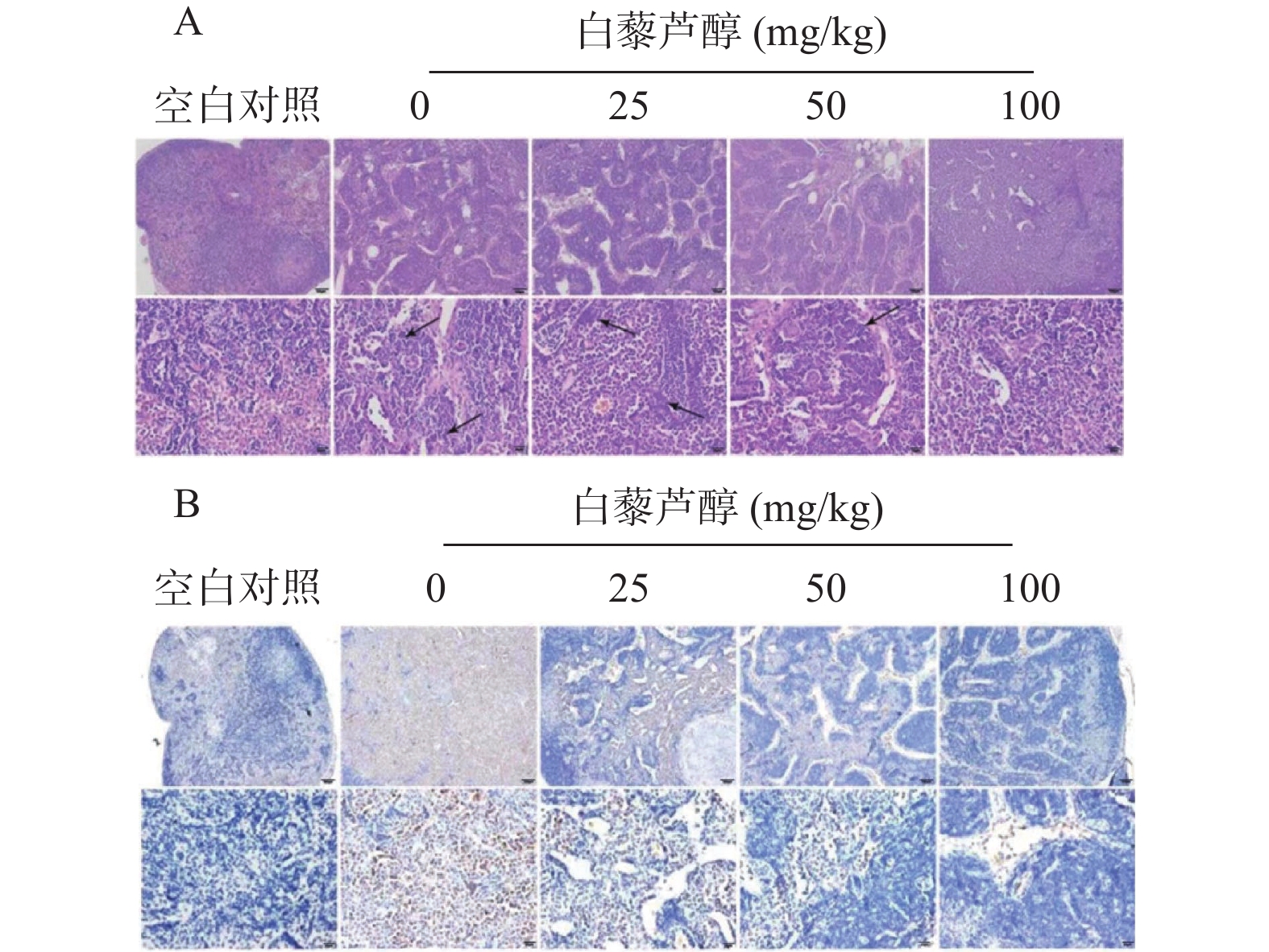
2.5 白藜芦醇可抑制体内鼻咽癌的转移能力
裸鼠淋巴结检测发现,见图5,足掌皮下成瘤后,瘤侧腘窝淋巴结与空白对照组相比明显肿大。给予白藜芦醇灌胃治疗后,随白藜芦醇浓度增高,淋巴结肿大程度明显减轻。实验结果证明,白藜芦醇灌胃能明显减轻瘤侧腘窝淋巴结的肿大程度。
HE染色发现(图6A):空白对照组(正常淋巴结)细胞排列规则,形态正常;足掌皮下成瘤后,未给予白藜芦醇治疗组,同侧淋巴结内出现多个转移灶(箭头所示,下同),淋巴结正常组织结构破坏,肿瘤细胞排列紊乱,聚集成堆,细胞边界不清或呈一群不规则裸核;给予白藜芦醇治疗组,随浓度由低到高,转移灶减少,淋巴结结构渐趋正常。
免疫组化发现(图6B):空白对照组(正常淋巴结)未见Keratin18阳性细胞;足掌皮下成瘤后,未给予白藜芦醇治疗组,同侧淋巴结内出现大量Keratin18阳性细胞(棕褐色,代表转移的肿瘤细胞);给予白藜芦醇治疗组,随着浓度由低到高,Keratin18阳性细胞逐渐减少。组织学检测显示,白藜芦醇灌胃明显抑制了鼻咽癌细胞由足掌皮下成瘤部位向腘窝淋巴结的转移,且抑制作用呈剂量依赖性。
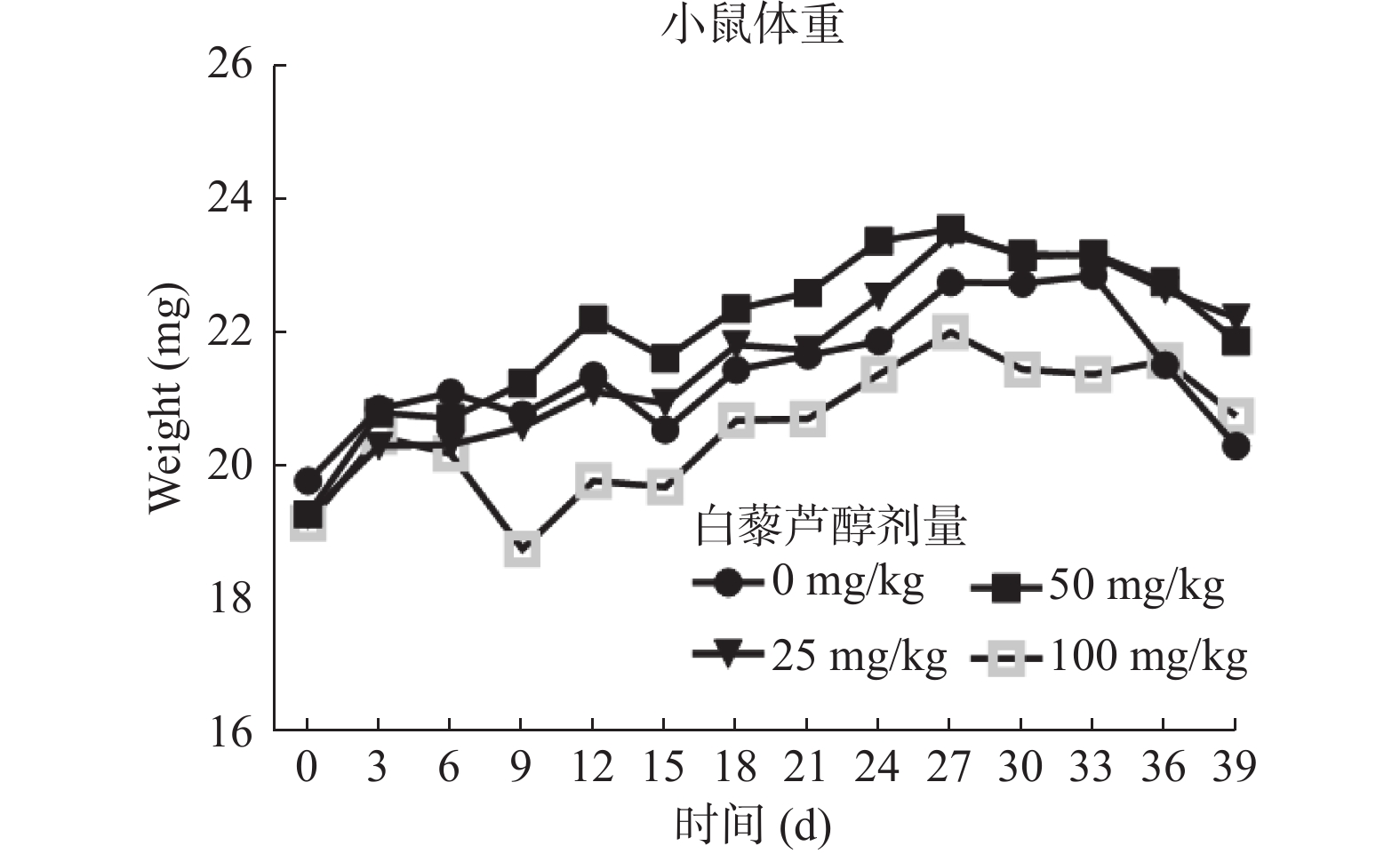
2.6 白藜芦醇灌胃治疗对荷瘤小鼠体重无明显影响
体重测量发现,在裸鼠足掌皮下注射鼻咽癌CNE2细胞后,每日经灌胃给予裸鼠不同剂量(0 mg/kg、25 mg/kg、50 mg/kg、100 mg/kg)的白藜芦醇,每3 d测量一次体重,各组裸鼠体重未见明显差异,见图7。
3. 讨论
中药白藜芦醇为天然多酚类化合物,广泛分布于虎杖、葡萄、花生等 70 余 种植物中,具有抗肿瘤、抗氧化、免疫调节等多种生物学效应。白藜芦醇可通过抑制 TGF-β、Wnt/β-Catenin、PI3K/Akt/NF-κB、Akt/GSK-3β/Snail 等信号通路增加上皮表型标志物E-钙黏蛋白(E-cadherin)和β-连环素(β-catenin)的表达、减少间质表型标志物N-钙粘蛋白(N-cadherin)、波形蛋白(Vimentin)和纤维连接蛋白(Fibronectin)的表达、抑制上皮间质转化(epithelial-mesenchymal transition,EMT),进而抑制口腔癌、乳腺癌、结直肠癌等恶性肿瘤的侵袭和转移[2-5]。白藜芦醇能否抑制鼻咽癌的转移能力,目前国内外研究报导较少。本研究Transwell实验发现:白藜芦醇能抑制鼻咽癌CNE2细胞的迁移、侵袭能力,在0至80 μmol/L浓度范围内,随着白藜芦醇浓度的增高,对鼻咽癌CNE2细胞迁移、侵袭能力的抑制作用逐渐增强。动物实验进一步证实了白藜芦醇能明显抑制鼻咽癌细胞由裸鼠足掌皮下成瘤部位向腘窝淋巴结的转移,其抑制作用呈浓度依赖性。
鼻咽癌的转移涉及到多种分子机制及信号通路,国内外研究发现,热休克蛋白27(hsp27)、埃兹蛋白(Ezrin)、核转录因子κB(NF-κB)、基质金属蛋白酶(MMPs)、上皮间质转化(EMT)等因子与鼻咽癌的转移密切相关[6-10]。白藜芦醇能否通过调节上述因子影响鼻咽癌的侵袭和迁移能力,目前国内外尚未见研究报导。
HSP27是热休克蛋白家族的主要成员之一,其重要的生物学功能是保护细胞免受各种应激因素的损伤,并参与调节细胞的增殖、分化及凋亡。磷酸化HSP27(p-HSP27)是HSP27的活性形式,其高表达与多种肿瘤如鼻咽癌、乳腺癌、前列腺癌、头颈鳞癌等的发生、发展、侵袭有关。HSP27是NF-κB的上游调节分子之一,研究发现:HSP27的过度表达可激活PI3K/AKT/NF-κB信号通路、诱导EMT,进而促进唾液腺样囊性癌、肺癌、肝癌等肿瘤的侵袭和迁移[11-14];此外,抑制Hsp27的表达可导致MMPs的活性明显下降,表明Hsp27对MMPs活性具有一定抑制作用[15]。本研究显示:白藜芦醇处理鼻咽癌CNE2细胞对总HSP27的表达无明显影响,但对磷酸化Hsp27的表达有明显抑制作用,表明白藜芦醇可通过抑制Hsp27的磷酸化进而抑制鼻咽癌的侵袭和转移能力。
埃兹蛋白(Ezrin)为连接于细胞膜和细胞骨架之间的埃兹蛋白-根蛋白-膜突蛋白 (Ezrin-radixin-moesin,ERM)家族成员之一,主要表达于各种类型的上皮细胞,具有调节细胞形态、粘附和活性,稳定细胞间连接,参与信号转导等功能。研究发现,磷酸化的Ezrin(p-Ezrin)可通过活化NF-κB信号通路促进结直肠癌的转移,其可能是NF-κB的上游调节因子; Ezrin也可通过调节E-cadherin、整合素促进鼻咽癌转移,还可通过降解NGX6a 、TMEM8B-a增加鼻咽癌的侵袭和转移能力[16, 17, 18]。本研究中,白藜芦醇处理鼻咽癌CNE2细胞对Ezrin总量无明显影响,但能抑制Ezrin的磷酸化水平,提示白藜芦醇可以通过抑制Ezrin的活化进而抑制鼻咽癌的转移能力。
核转录因子κB(NF-κB)是一种作用十分广泛的真核细胞转录因子,正常情况下,NF-κB与其抑制蛋白IĸB结合,以无活性的复合物形式存在于胞浆中。在某些细胞外信号(如细菌,病毒等)或致瘤因素的刺激下,IĸB 激酶(IKK)活化,引起IĸB-ɑ的丝氨酸残基磷酸化,随后泛素化降解,释放出NF-κB,NF-κB由细胞质转入细胞核,与特定的DNA系列结合,启动相关基因的转录,参与调控细胞的增生、分化和凋亡。IĸB-ɑ是hsp27的靶蛋白之一,hsp27可促进IĸB-ɑ的泛素化和后续降解,从而增强NF-ĸB的活性。研究发现:NF-ĸB的异常活化和表达与肿瘤的侵袭和转移密切相关[19] ,NF-κB信号通路的激活是MMP2、MMP9表达所必须的;多种因子可通过NF-κB信号通路促进EMT、增加MMP的表达水平,进而促进鼻咽癌的转移能力[7-8, 20-21];白藜芦醇可通过抑制NF-κB的转录活性抑制MMP的表达、进而抑制恶性胶质瘤、肺癌等肿瘤的转移[22-23]。本研究中,免疫荧光实验初步证实,白藜芦醇可能抑制CNE2细胞中NF-κB/p65的核转位,从而降低NF-κB的转录活性,抑制NF-κB下游转移相关靶基因的表达,与其他学者所发现的白藜芦醇对肿瘤细胞中NF-κB的抑制效应一致,后续课题组将进一步通过EMSA实验观察白藜芦醇对NF-κB核转位的抑制作用。
EMT是指某些生理和病理状态下出现的上皮细胞向间质细胞转化的现象,表现为上皮表型标志物 E-cadherin 和β-catenin的表达减少,间质表型标志物N-cadherin、 Vimentin和Fibronectin的表达增加,是上皮源性恶性肿瘤侵袭和转移的关键步骤之一。MMPs是一个大的蛋白质家族,其能通过降解细胞外基质破坏肿瘤细胞侵袭转移的组织学屏障,在肿瘤的侵袭和转移中起关键作用,是肿瘤侵袭转移过程中的主要蛋白水解酶,目前研究较多的是MMP-2、MMP-9。研究发现EMT及MMP在鼻咽癌的转移中发挥重要作用[9, 21, 24-25]。本研究发现:不同浓度白藜芦醇处理鼻咽癌CNE2细胞24 h后,上皮标志物E-cadherin的表达水平上升,但间质标志物Vimentin水平无明显变化,提示白藜芦醇可通过上调E-cadherin抑制CNE2细胞的上皮间质转化,从而降低CNE2细胞的变形迁移能力;对于Vimentin水平无明显变化的原因尚需进一步研究。本研究用Western blot实验检测了MMPs蛋白的表达水平,结果显示,白藜芦醇对MMP2和MMP9均有较明显的抑制作用,由此可见,白藜芦醇能通过抑制MMP2和MMP9的表达进而抑制鼻咽癌的侵袭迁移能力。
综上所述,本研究证实白藜芦醇在体外和体内均能有效抑制鼻咽癌CNE2细胞的迁移、侵袭和转移,其分子机制可能与肿瘤转移相关蛋白Ezrin和Hsp27的磷酸化水平下调、NF-κB/p65核转位抑制、EMT相关蛋白E-cadherin表达上调、基质金属蛋白酶家族MMP2和MMP9下调密切相关。白藜芦醇可能通过抑制Ezrin、Hsp27的磷酸化,进而抑制NF-κB/p65的核转位,从而下调MMP2、MMP9 的表达水平、上调E-cadherin的水平进而抑制EMT,最终抑制鼻咽癌的转移能力。
-
表 1 所选SNP位点信息
Table 1. The information of selected SNPs in the current study
SNPs 基因 功能 位置 等位基因 中国南方汉族人群MAF rs1348808 HOXD-AS2 启动子区域 Chr 2: 176135888 C>T 0.28 rs2431099 MIR3142HG 增强子区域 Chr 5: 160459613 A>G 0.33 表 2 2个SNP位点在3组中等位基因及基因型分布频率结果 [n (%)]
Table 2. Allele and genotype frequencies of two SNPs between three groups [n(%)]
SNPs 等位基因/基因型 对照组 CIN2组 CIN3组 χ2 P rs1348808 C 529(27.1) 34(33.3) 164(22.3) 9.348 0.009* T 1423 (72.9)68(66.7) 572(77.7) C/C 78(8.0) 3(5.9) 22(6.0) 13.525 0.009* C/T 373(38.2) 28(54.9) 120(32.6) T/T 525(53.8) 20(39.2) 226(61.4) rs2431099 A 720(36.9) 23(22.5) 254(34.5) 9.327 0.009* G 1232 (63.1)79(77.5) 482(65.5) A/A 136(13.9) 6(11.8) 36(9.8) 19.513 0.001* A/G 448(45.9) 11(21.5) 182(49.4) G/G 392(40.2) 34(66.7) 150(40.8) *P < 0.016(经Bonferroni校正,n = 3)。 表 3 rs1348808位点遗传模式分析
Table 3. The inheritance model analysis of rs1348808
模型 CIN3 vs 对照组 CIN3 vs CIN2 OR (95%CI) P AIC BIC OR (95%CI) P AIC BIC 共显性 T/T 1.00 0.037 1577.3 1592.9 1.00 0.008 306.6 318.7 C/T 0.75(0.58~0.97) 2.63(1.43~4.76) C/C 0.65(0.40~1.08) 1.54(0.42~5.56) 显性 T/T 1.00 0.012 1575.5 1586.0 1.00 0.003 305.4 313.5 C/T-C/C 0.73(0.57~0.93) 2.44(1.35~4.55) 隐性 T/T-C/T 1.00 0.200 1580.2 1590.7 1.00 0.980 314.3 322.4 C/C 0.73(0.45~1.19) 0.98(0.28~3.45) 超显性 T/T-C/C 1.00 0.056 1578.2 1588.6 1.00 0.002* 305.0 313.1 C/T 0.78(0.61~1.01) 2.50(1.39~4.55) 逻辑累加 --- 0.78(0.64~0.95) 0.012* 1575.5 1585.9 1.72(1.11~2.70) 0.018 308.8 316.9 *P < 0.05;且AIC和BIC值最小,为分组比较中的最优遗传模式。 表 4 rs2431099位点在CIN2组和对照组的比较中遗传模式分析
Table 4. The inheritance model analysis of rs2431099 between the CIN2 and control groups
模型 OR (95%CI) P AIC BIC 共显性 G/G 1 5.00×10−4 396.7 411.5 A/G 0.28(0.14~0.56) A/A 0.51(0.21~1.23) 显性 G/G 1 2.00×10−4* 395.9 405.7 A/G-A/A 0.34(0.18~0.61) 隐性 G/G-A/G 1 0.66 409.5 419.4 A/A 0.83(0.34~1.96) 超显性 G/G-A/A 1 4.00×10−4 397.2 407.1 A/G 0.32(0.16~0.64) 逻辑累加 --- 0.51(0.32~0.81) 0.003 400.7 410.6 *P < 0.05;且AIC和BIC值最小,为分组比较中的最优遗传模式。 -
[1] Arbyn M,Ronco G,Anttila A,et al. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer[J]. Vaccine,2012,30(Suppl 5):F88-F99. [2] 赵超,毕蕙,赵昀,等. 子宫颈高级别上皮内病变管理的中国专家共识[J]. 中国妇产科临床杂志,2022,23(2):220-224. [3] Mccredie M R,Sharples K J,Paul C,et al. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: A retrospective cohort study[J]. Lancet Oncol,2008,9(5):425-434. doi: 10.1016/S1470-2045(08)70103-7 [4] Zheng R,Zhang S,Zeng H,et al. Cancer incidence and mortality in China,2016[J]. Journal of the National Cancer Center,2022,2(1):1-9. doi: 10.1016/j.jncc.2022.02.002 [5] Kawase K,Taguchi A,Ishizaka A,et al. Allelic loss of HLA class I facilitates evasion from immune surveillance in cervical intraepithelial neoplasia[J]. HLA,2024,103(6):e15509. doi: 10.1111/tan.15509 [6] Hangauer M J,Vaughn I W,Mcmanus M T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs[J]. PLoS Genet,2013,9(6):e1003569. doi: 10.1371/journal.pgen.1003569 [7] Kaewsapsak P,Shechner D M,Mallard W,et al. Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking[J]. Elife,2017,6(1):e29224. [8] Melé M,RInn J L. "Cat's cradling" the 3D genome by the act of lncRNA transcription[J]. Mol Cell,2016,62(5):657-664. doi: 10.1016/j.molcel.2016.05.011 [9] Lee S,Kopp F,Chang T C,et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins[J]. Cell,2016,164(1-2):69-80. doi: 10.1016/j.cell.2015.12.017 [10] Benoit bouvrette L P,Cody N A L,Bergalet J,et al. CeFra-seq reveals broad asymmetric mRNA and noncoding RNA distribution profiles in Drosophila and human cells[J]. RNA,2018,24(1):98-113. doi: 10.1261/rna.063172.117 [11] Dai J,Zhang S,Shi Y,et al. rs217727 of lncRNA H19 is associated with cervical cancer risk in the Chinese Han population[J]. Pharmgenomics Pers Med,2023,16(1):933-948. [12] Liu Y,Zhang Q,Ni R. Association between genetic variants (rs920778,rs4759314,and rs217727) in LncRNAs and cervical cancer susceptibility in Chinese population: A systematic review and meta-analysis[J]. Front Genet,2022,13(1):988207. [13] 中华医学会. 临床诊疗指南: 妇产科学分册 [M]. 北京: 人民卫生出版社,2007: 65-67. [14] Yang J,Yan Z,Wang Y,et al. Association study of relationships of polymorphisms in the miR-21,miR-26b,miR-221/222 and miR-126 genes with cervical intraepithelial neoplasia and cervical cancer[J]. BMC Cancer,2021,21(1):997. doi: 10.1186/s12885-021-08743-2 [15] Shi Y Y,He L. SHEsis,a powerful software platform for analyses of linkage disequilibrium,haplotype construction,and genetic association at polymorphism loci[J]. Cell Res,2005,15(2):97-98. doi: 10.1038/sj.cr.7290272 [16] Solé X,Guinó E,Valls J,et al. SNPStats: A web tool for the analysis of association studies[J]. Bioinformatics,2006,22(15):1928-1929. doi: 10.1093/bioinformatics/btl268 [17] Qi Y,Wang Z,Wu F,et al. Long noncoding RNA HOXD-AS2 regulates cell cycle to promote glioma progression[J]. J Cell Biochem,2019,120(5):8343-8351. doi: 10.1002/jcb.28117 [18] Zhang Y,Ma H. LncRNA HOXD-AS2 regulates miR-3681-5p/DCP1A axis to promote the progression of non-small cell lung cancer[J]. J Thorac Dis,2023,15(3):1289-1301. doi: 10.21037/jtd-23-153 [19] Paterson M R,Kriegel A J. MiR-146a/b: A family with shared seeds and different roots[J]. Physiol Genomics,2017,49(4):243-252. doi: 10.1152/physiolgenomics.00133.2016 [20] Guo X,Zhang M,Li Q,et al. Evaluation of genetic variants in MIR3142HG in susceptibility to and prognosis of glioma[J]. Am J Clin Oncol,2020,43(1):1-8. doi: 10.1097/COC.0000000000000587 [21] Chen Y,Zhao Y,Lu R,et al. Identification and validation of a novel genomic instability-associated long non-coding RNA prognostic signature in head and neck squamous cell carcinoma[J]. Front Cell Dev Biol,2021,9(1):787766. [22] Järvelin A I,Noerenberg M,Davis I,et al. The new (dis)order in RNA regulation[J]. Cell Commun Signal,2016,14(1):9. doi: 10.1186/s12964-016-0132-3 [23] Hentze M W,Castello A,Schwarzl T,et al. A brave new world of RNA-binding proteins[J]. Nat Rev Mol Cell Biol,2018,19(5):327-341. doi: 10.1038/nrm.2017.130 期刊类型引用(0)
其他类型引用(1)
-






 下载:
下载:




 下载:
下载:

