Characteristics of Intestinal Microflora in Patients with Coronary Heart Disease
-
摘要:
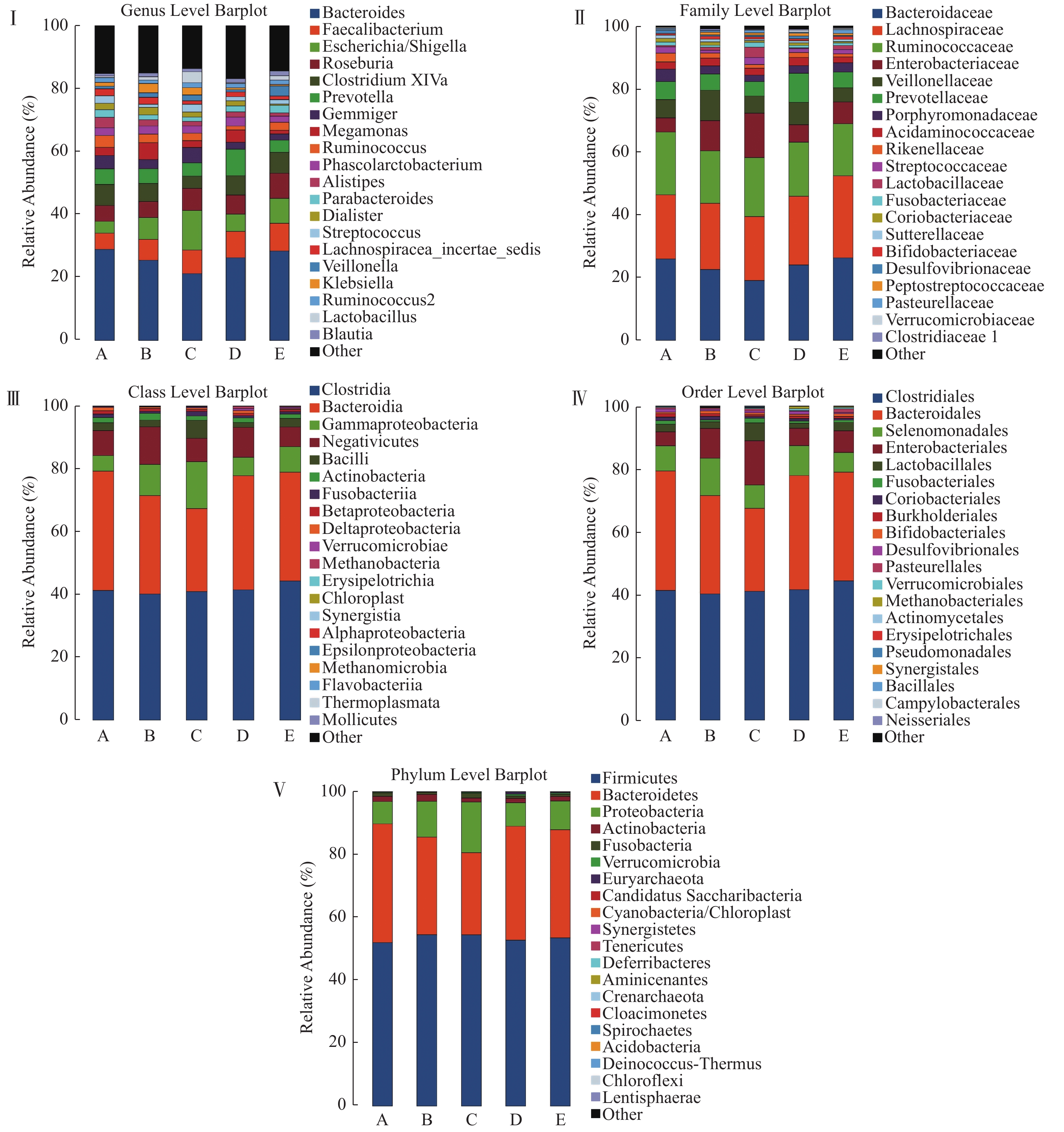
目的 分析云南冠心病人群肠道菌群结构和多样性特征,探究不同类型冠心病人群之间,及与无冠脉病变人群肠道菌群结构差异。 方法 共采集急性心肌梗死患者67名(A组)、不稳定型心绞痛患者91名(B组)、稳定型心绞痛患者47名(C组)、冠脉无异常者64名(D组)、冠状动脉粥样硬化患者37名(E组),提取粪便DNA,根据细菌16srRNA v3-v4序列设计特异性引物,进行PCR扩增,使用Illumina HiSeq PE250进行测序,测序结果进行物种组成统计,Alpha多样性,组间差异分析,最终得到样品物种信息。 结果 各组间肠道菌群结构存在差异(P < 0.05)。与D组相比,A组的梭杆菌增多,B组的拟杆菌减少,变形杆菌、放线菌增多,C组的拟杆菌减少,变形杆菌、梭杆菌增多,E组变形杆菌增多( P < 0.05)。Alpha多样性发现显示急性心肌梗死患者组的Alpha多样性高于其他组( P < 0.05)。 结论 冠状动脉粥样硬化患者、稳定型心绞痛患者、不稳定型心绞痛患者、急性心肌梗死患者与对照组肠道菌群的结构、多样性不同,他们存在不同程度的肠道菌群失调。 Abstract:Objective To analyze the composition and diversity of intestinal microflora in patients with coronary heart disease in Yunnan, and to explore the differences of intestinal microflora among different types of coronary heart disease and those without coronary artery disease. Methods 67 patients with acute myocardial infarction(group A), 91 patients with unstable angina(group B), 47 patients with stable angina(group C), 64 patients without abnormal coronary arteries(group D)and 37 patients with coronary atherosclerosis(group E)were enrolled. Faeces sample DNA was extracted. Specific primers were designed according to 16SrRNA v3-v4 sequence of bacteria, PCR amplification was carried out, and sequenced with Illumina hiseq PE250. The sequencing results were analyzed by species composition statistics, Alpha diversity, intergroup difference analysis, and finally get the species information of the faeces sample. Results There were differences in the composition of intestinal flora among groups(P < 0.05). Compared with group D, there were more Clostridium in group A, less Bacteroides, more Proteus and actinomycetes in group B, less Bacteroides, more Proteus and Clostridium in group C, and more Proteus in group E( P < 0.05). The alpha diversity of AMI patients was higher than that of other groups( P < 0.05). Conclusion The composition, diversity and abundance of intestinal flora in coronary atherosclerotic group, stable angina group, unstable angina group, acute myocardial infarction group and control group were different in Yunnan, and they had intestinal flora disorder at various degrees. -
Key words:
- Coronary artery disease /
- Atherosclerosis /
- Intestinal microorganism /
- 16srRNA
-
表 1 五组间一般临床检测资料
$ \left( {{{\bar x}} \pm {{s}}} \right)$ Table 1. General clinical examination data of five groups
$ \left( {{{\bar x}} \pm {{s}}} \right)$ 组别 男性
n(%)年龄
(岁)BMI
(kg/m2)空腹血糖
(mmol/L)cTnI TG
(mmol/L)LDL-C
(mmol/L)ALT
(U/L)Cr
(μmol/L)高血压
n(%)糖尿病
n(%)A 61(91.0) 57.13 ± 12.36 24.94 ± 3.01 6.55 ± 2.74 17.17 ± 24.89 1.80 ± 0.11 2.83 ± 0.77 40.15 ± 20.61 71.12 ± 24.98 31(46.3)# 12(17.9) B 48(60.0)* 62.43 ± 9.95 24.79 ± 3.40 6.32 ± 2.81 0.01 ± 0.06* 1.94 ± 1.23 2.55 ± 1.21 29.84 ± 14.53 65.39 ± 15.79 50(62.5)# 19(23.8) C 23(52.3)* 58.43 ± 9.69 24.34 ± 3.46 5.96 ± 2.37 0.01 ± 0.029* 1.78 ± 0.91 2.78 ± 0.78 27.76 ± 14.39* 68.00 ± 18.41 19(43.2)# 11(25.0) D 18(35.3)* 53.92 ± 12.63∆ 23.52 ± 3.32 5.56 ± 1.33 0.00 ± 0.00* 1.49 ± 0.73 2.65 ± 0.86 26,48 ± 15.04* 60.33 ± 13.52* 17(33.3)#∆ 6(11.8) E 18(62.1)* 63.86 ± 8.72▲ 24.48 ± 3.31 5.90 ± 2.31 0.00 ± 0.00* 1.69 ± 0.89 2.42 ± 0.88 27.79 ± 17.93* 66.29 ± 14.31 21(72.4)* 6(20.7) 与A组相比,*P < 0.05;与E组相比 #P < 0.05;与B组相比 ∆P < 0.05;与D组相比 ▲P < 0.05。 表 2 差异显著物种列表
Table 2. List of species with significant difference(Genus Level)
物种名称 A组(均值) B组(均值) C组(均值) D组(均值) E组(均值) P g__Acinetobacter 5.31E-05 3.17E-04 1.04E-03 1.03E-05 1.56E-05 0.01 g__Akkermansia 2.68E-03 5.39E-04 2.48E-03 5.92E-03 3.61E-03 4.51E-04 g__Alistipes 0.03 0.02 0.01 0.01 9.33E-03 2.39E-05 g__Anaerofustis 0 0 1.76E-06 0 4.47E-06 0.04 g__Anaerotruncus 1.58E-04 9.54E-05 7.74E-05 3.20E-04 1.81E-04 2.85E-03 g__Atopobium 2.07E-04 6.63E-05 2.99E-04 3.62E-05 4.69E-05 9.73E-04 g__Bilophila 4.71E-03 1.75E-03 1.25E-03 5.45E-03 1.28E-03 3.09E-05 g__Clostridium XI 1.82E-03 8.87E-03 7.20E-03 8.41E-03 5.92E-03 2.42E-03 g__Clostridium XVIII 1.37E-04 1.71E-04 1.00E-03 1.96E-04 2.79E-04 4.56E-03 g__Clostridium XlVa 0.06 0.05 0.03 0.05 0.06 0.01 g__Clostridium XlVb 6.47E-03 5.16E-03 2.48E-03 5.47E-03 2.86E-03 3.44E-03 g__Clostridium sensu stricto 1.75E-03 5.10E-03 1.99E-03 3.60E-03 1.96E-03 0.02 g__Cryptobacterium 1.60E-05 2.73E-06 0 1.29E-06 4.47E-06 9.27E-03 g__Desulfovibrio 3.03E-03 1.29E-03 1.48E-03 1.15E-03 8.05E-04 0.02 g__Eikenella 1.23E-06 0 8.80E-06 0 2.23E-06 8.07E-03 g__Eisenbergiella 2.02E-03 1.12E-03 4.29E-04 7.34E-04 8.80E-04 0.01 g__Flavobacterium 0 0 1.23E-05 0 0 0.03 g__Gemella 1.10E-04 9.36E-05 2.29E-04 1.07E-04 5.14E-05 0.03 g__Gemmiger 0.03 0.02 0.04 0.02 0.02 0.04 g__Granulicatella 3.01E-04 1.90E-04 4.47E-04 1.34E-04 1.56E-04 8.03E-03 g__Klebsiella 0.01 0.03 0.02 5.33E-03 3.84E-03 1.92E-03 g__Lactobacillus 4.16E-03 9.76E-03 0.03 3.07E-03 0.01 4.18E-03 g__Megamonas 0.02 0.05 0.02 0.04 0.01 0.01 g__Odoribacter 2.34E-03 1.66E-03 5.49E-04 1.25E-03 7.04E-04 4.06E-04 g__Oscillibacter 3.73E-03 3.04E-03 1.38E-03 2.57E-03 1.45E-03 2.58E-04 g__Parabacteroides 0.02 0.01 0.01 0.02 0.02 1.65E-04 g__Paraprevotella 7.95E-03 5.86E-03 3.33E-03 6.34E-03 3.21E-03 0.05 g__Porphyromonas 6.66E-05 5.27E-05 6.51E-05 2.33E-05 4.92E-05 0.02 g__Propionibacterium 3.70E-06 9.99E-06 8.80E-06 1.29E-06 0 0.04 g__Rothia 2.11E-04 7.00E-05 1.39E-04 7.23E-05 1.85E-04 0.01 g__Ruminococcus 0.03 0.02 0.02 0.01 0.02 0.02 g__Streptococcus 0.02 8.37E-03 0.02 0.01 0.01 2.95E-05 g__Veillonella 6.73E-03 0.01 0.02 5.63E-03 0.03 8.24E-04 -
[1] Gill S R,Pop M,Deboy R T,et al. Metagenomic analysis of the human distal gut microbiome[J]. Science,2006,312(5778):1355-1359. doi: 10.1126/science.1124234 [2] Qin J,Li R,Raes J,et al. A human gut microbial gene catalogue established by metagenomic sequencing[J]. Nature,2010,464(7285):59-65. doi: 10.1038/nature08821 [3] Nicholson J K,Holmes E,Wilson I D. Gut microorganisms,mammalian metabolism and personalized health care[J]. Nature Reviews Microbiology,2005,3(5):431-438. doi: 10.1038/nrmicro1152 [4] Perry R J,Peng L,Barry N A,et al. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome[J]. Nature,2016,534(7606):213-217. doi: 10.1038/nature18309 [5] Pedersen H K,Gudmundsdottir V,Nielsen H B,et al. Human gut microbes impact host serum metabolome and insulin sensitivity[J]. Nature,2016,535(7612):376-381. doi: 10.1038/nature18646 [6] Komaroff A L. The microbiome and risk for obesity and diabetes[J]. JAMA,2017,317(4):355-356. doi: 10.1001/jama.2016.20099 [7] Chu H,Khosravi A,Kusumawardhani I P,et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease[J]. Science,2016,352(6289):1116-1120. doi: 10.1126/science.aad9948 [8] Man S M,Zhu Q,Zhu L,et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer[J]. Cell,2015,162(1):45-58. doi: 10.1016/j.cell.2015.06.001 [9] Buffington S A,Di Prisco G V,Auchtung T A,et al. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring[J]. Cell,2016,165(7):1762-1775. doi: 10.1016/j.cell.2016.06.001 [10] Akbari E,Asemi Z,Daneshvar Kakhaki R,et al. Effect of probiotic supplementation on cognitive function and metabolic status in alzheimer's disease:A randomized,double-blind and controlled trial[J]. Front Aging Neurosci,2016,8:256. [11] 陈晓晴,王南,荆志成,等. 肠道菌群与心血管疾病研究进展[J].中国实用内科杂志,2018,38(8):766-769. [12] Liu H,Chen X,Hu X,et al. Alterations in the gut microbiome and metabolism with coronary artery disease severity[J]. Microbiome,2019,7(1):68. doi: 10.1186/s40168-019-0683-9 [13] 郭云双,袁宇. 肠道菌群与豫北地区心血管疾病的关系及其作用机制探讨[J].河南医学研究,2018,27(12):2115-2118. doi: 10.3969/j.issn.1004-437X.2018.12.002 [14] Deschasaux M,Bouter K E,Prodan A,et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography[J]. Nat Med,2018,24(10):1526-1531. doi: 10.1038/s41591-018-0160-1 [15] Emoto T,Yamashita T,Sasaki N,et al. Analysis of gut microbiota in coronary artery disease patients:A possible link between gut microbiota and coronary artery disease.[J]. Journal of Atherosclerosis & Thrombosis,2016,23(8):908. [16] Mazmanian S K,Round J L,Kasper D L. A microbial symbiosis factor prevents intestinal inflammatory disease[J]. Nature,2008,453(7195):620-625. doi: 10.1038/nature07008 [17] Wu G D,Chen J,Hoffmann C,et al. Linking long-term dietary patterns with gut microbial enterotypes[J]. Science,2011,334(6052):105-108. doi: 10.1126/science.1208344 [18] Kappel B A,Federici M. Gut microbiome and cardiometabolic risk[J]. Rev Endocr Metab Disord,2019,20(4):399-406. doi: 10.1007/s11154-019-09533-9 [19] Xu H,Wang X,Feng W,et al. The gut microbiota and its interactions with cardiovascular disease[J]. Microb Biotechnol,2020,13(3):637-656. doi: 10.1111/1751-7915.13524 [20] Zhang S,Yu N,Arce R M. Periodontal inflammation:Integrating genes and dysbiosis[J]. Periodontol,2020,82(1):129-142. doi: 10.1111/prd.12267 [21] Carrizales-Sepúlveda E F,Ordaz-Farías A,Vera-Pineda R,et al. Periodontal disease,systemic inflammation and the risk of cardiovascular disease[J]. Heart Lung Circ,2018,27(11):1327-1334. doi: 10.1016/j.hlc.2018.05.102 [22] Cross B,Faustoferri R C,Quivey R G Jr. What are we learning and what can we learn from the human oral microbiome project?[J]. Curr Oral Health Rep,2016,3(1):56-63. doi: 10.1007/s40496-016-0080-4 [23] 李俊艳,孙致远,袁宇. 基于高通道测序的肠道菌群与冠心病的相关性研究[J].中国全科医学,2019,22(29):3554-3560. [24] He Y,Wu W,Zheng H M,et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models[J]. Nat Med,2018,24(10):1532-1535. doi: 10.1038/s41591-018-0164-x -






 下载:
下载: