Analysis of lncRNA-mRNA Co-expression Network in Invasive Pituitary Adenomas
-
摘要:
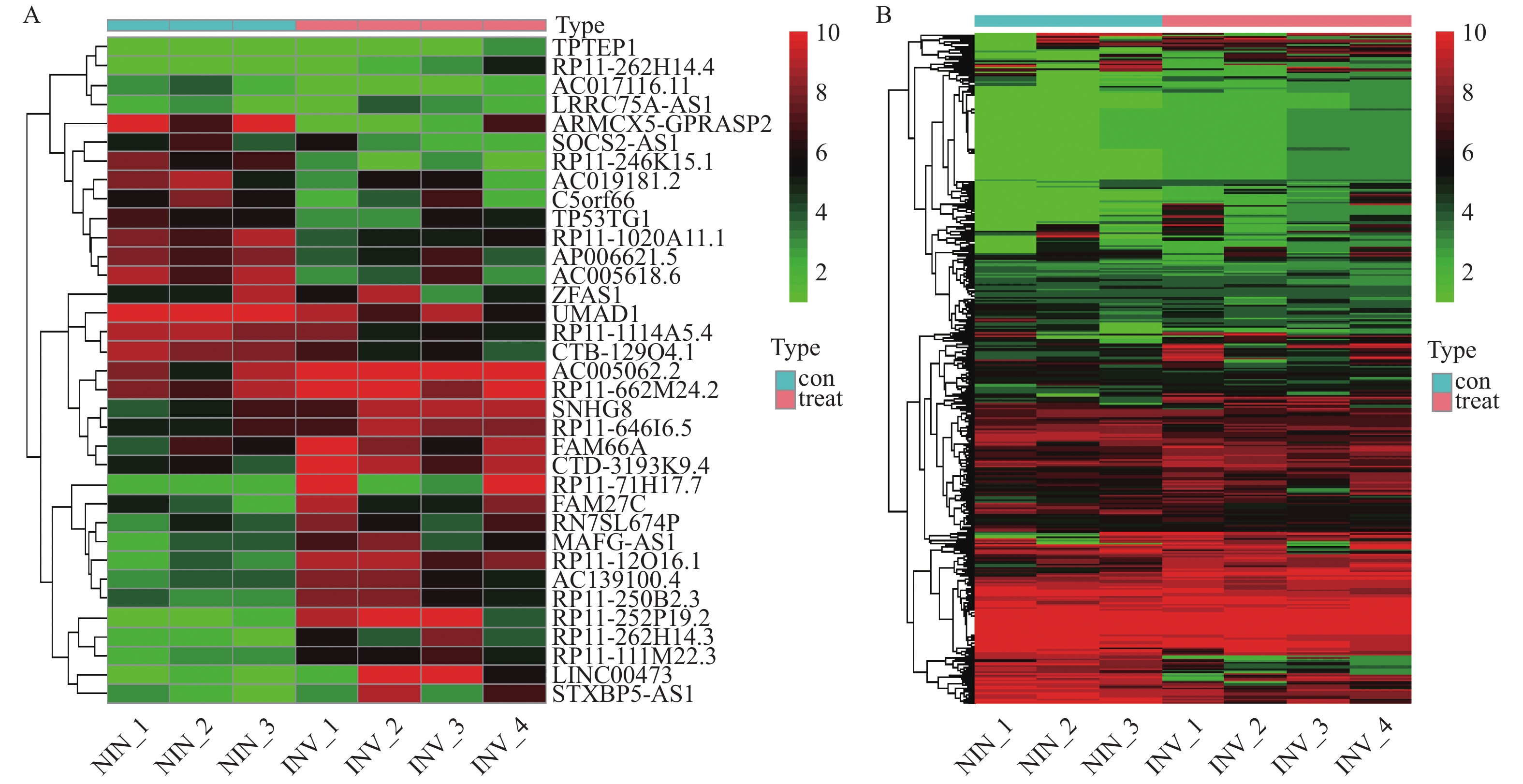
目的 利用二代测序技术筛选侵袭性垂体腺瘤组织中差异表达的lncRNA,分析其在侵袭性垂体腺瘤发病机制中可能发挥的潜在作用。 方法 通过RNA测序分析4例侵袭性垂体腺瘤患者和3例非侵袭性垂体腺瘤患者的lncRNA和mRNA的表达情况,筛选出差异表达的lncRNA和mRNA。据此,构建lncRNA-mRNA共表达网络,并进行GO和KEGG通路分析。 结果 对侵袭性和非侵袭性垂体瘤的lncRNA和mRNA差异表达分析,共获得35个差异表达的lncRNA和463个差异表达的mRNA。共表达网络显示FAM66A、TPTEP1、AC017116.11、RP11-1114A5.4和RP11-246K15.1在网络中处于关键位置。 结论 lncRNA-mRNA共表达网络的建立,发现一些差异表达的lncRNA可能在侵袭性垂体腺瘤的发病机制中发挥关键作用,可能成为侵袭性垂体腺瘤新的诊断标志物和治疗靶点。 -
关键词:
- 侵袭性垂体腺瘤 /
- 长链非编码(lncRNA) /
- RNA测序 /
- lncRNA-mRNA共表达
Abstract:Objective To screen differentially expressed lncRNAs in invasive pituitary adenomas by second-generation sequencing, and to explore their potential role in the pathogenesis of invasive pituitary adenomas. Methods The expressions of lncRNA and mRNA in 4 patients with invasive pituitary adenoma and 3 patients with non-invasive pituitary adenoma were analyzed by RNA sequencing to single out the differentially expressed lncRNA and mRNA. Accordingly, lncrNA-mRNA co-expression networks were constructed and GO and KEGG pathways were analyzed. Results A total of 35 differentially expressed lncRNAs and 463 differentially expressed mRNAs were obtained by analyzing the differentially expressed lncRNAs and mRNAs in both invasive and non-invasive pituitary tumors. The co-expression network showed that FAM66A, TPTEP1, AC017116.11, RP11-1114A5.4 and RP11-246K15.1 were in a key position in the network. Conclusion Through the establishment of co-expression network of lncRNA-mRNA, we found that some new lncRNA may play a key role in the pathogenesis of invasive pituitary adenomas and may be used as novel biomarkers of invasive pituitary adenomas. -
表 1 差异表达的lncRNA
Table 1. Differentially expressed lncRNAs
基因名 Log2(Foldchange) P 异常表达情况 RP11-252P19.2 7.110230855 0.025594461 上调 LINC00473 6.892052835 0.000316802 上调 STXBP5-AS1 5.640589233 0.007878966 上调 RP11-71H17.7 5.221628067 0.017764884 上调 TPTEP1 4.89206949 0.003097546 上调 RP11-262H14.3 3.952787834 0.03606433 上调 RP11-12O16.1 2.807343125 0.011324588 上调 RP11-111M22.3 2.28836899 0.002792906 上调 RP11-250B2.3 1.906609127 0.035774432 上调 FAM66A 1.848133787 0.002209197 上调 RP11-262H14.4 1.813382724 0.009868508 上调 CTD-3193K9.4 1.721455664 0.001307362 上调 MAFG-AS1 1.536359193 0.044840975 上调 SNHG8 1.461730762 0.049804872 上调 AC139100.4 1.448409941 0.045905713 上调 FAM27C 1.415494365 0.029287703 上调 AC005062.2 1.349936987 0.033475711 上调 RP11-646I6.5 1.272730756 0.046225249 上调 RN7SL674P 1.260917811 0.008860604 上调 RP11-662M24.2 1.188403274 0.037409712 上调 LRRC75A-AS1 1.042079322 0.002182732 上调 ZFAS1 −1.030295183 0.02204746 下调 TP53TG1 −1.093552638 0.027463784 下调 CTB-129O4.1 −1.167587296 0.041920971 下调 RP11-1020A11.1 −1.227419109 0.025518005 下调 AP006621.5 −1.265098387 0.004293786 下调 C5orf66 −1.323699343 0.024003471 下调 RP11-1114A5.4 −1.346165585 0.036015113 下调 AC017116.11 −1.355620383 0.018311217 下调 AC019181.2 −1.556768999 0.031834266 下调 UMAD1 −2.049223764 0.026775377 下调 AC005618.6 −2.103056949 0.012754031 下调 SOCS2-AS1 −2.450904018 0.010320293 下调 RP11-246K15.1 −3.394395932 0.006322703 下调 ARMCX5-GPRASP2 −3.509890448 0.042606052 下调 表 2 排名前20的差异表达的mRNA
Table 2. The top 20 differentially expressed mRNA
mRNAs基因名 Log2(Foldchange) P 异常表达情况 SLC4A3 3.130642688 0.000067700 上调 ZNF444 1.555389418 0.000111993 上调 HDAC4 2.871048277 0.000146683 上调 MRO −3.344017691 0.000209461 下调 ECHDC1 10.92744203 0.000217674 上调 KIAA1958 −1.739496332 0.000224985 下调 TBC1D5 22.81214317 0.000254824 上调 NCALD 2.281098096 0.000325798 上调 LAMTOR5 −4.230692295 0.000366157 下调 TMPRSS6 7.265115275 0.000399492 上调 RPL26 −4.965389762 0.000462772 下调 RPSA 2.332207622 0.000464748 上调 CTSC 1.469599612 0.000464889 上调 MYO10 4.166108193 0.000472723 上调 ATP6AP2 3.197467642 0.000514505 上调 EPHX2 2.326018659 0.000616419 上调 PARP11 −1.579039067 0.000625344 下调 AGTRAP 1.045079801 0.000845595 上调 MXRA7 −1.391168544 0.000967649 下调 SLC29A2 −9.596231979 0.001050552 下调 表 3 Degree排名前五的lncRNA
Table 3. The top 5 lncRNAs in Degree
基因名 Degree FAM66A 52 TPTEP1 42 AC017116.11 33 RP11-1114A5.4 29 RP11-246K15.1 28 -
[1] Ezzat S,Asa S L,Couldwell W T,et al. The prevalence of pituitary adenomas:A systematic review[J]. Cancer,2004,101(3):613-619. doi: 10.1002/cncr.20412 [2] Sav A,Rotondo F,Syro L V,et al. Invasive,atypical and aggressive pituitary adenomas and carcinomas[J]. Endocrinology and Metabolism Clinics of North America,2015,44(1):99-104. [3] 王任直. 神经外科学[M]. 2 版. 北京: 人民卫生出版社, 2002: 462 - 463. [4] Ponting C P,Oliver P L. Evolution and functions of long noncoding RNAs[J]. Cell,2009,136(4):629-641. doi: 10.1016/j.cell.2009.02.006 [5] Peschansky V J. Non-coding RNAs as direct and indirect modulators of epigenetic regulation[J]. Epigenetics,2014,9(1):3-12. doi: 10.4161/epi.27473 [6] Gutschner T. The hallmarks of cancer:A long non-coding RNA point of view[J]. RNA Biology,2012,9(6):703-719. doi: 10.4161/rna.20481 [7] Lu G,Duan J. Long-noncoding RNA IFNG-AS1 exerts oncogenic properties by interacting with epithelial splicing regulatory protein 2 (ESRP2) in pituitary adenomas[J]. Pathology,Research and Practice,2018,214(12):2054-2061. [8] Yu G,Li C,Xie W,et al. Long non-coding RNA C5orf66-AS1 is downregulated in pituitary null cell adenomas and is associated with their invasiveness[J]. Oncology Reports,2017,38(2):1140-1148. doi: 10.3892/or.2017.5739 [9] Zhou L,Chen J,Li Z,et al. Integrated profiling of microRNAs and mRNAs:microRNAs located on Xq273 associate with clear cell renal cell carcinoma[J]. PloS One,2010,5(12):e15224. doi: 10.1371/journal.pone.0015224 [10] Chen L, Zhang Y H, Wang S, et al. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways[J]. PLoS One,2017,12(9):e0184129. [11] Chen C M,Lu Y L,Sio C P,et al. Gene Ontology based housekeeping gene selection for RNA-seq normalization[J]. Methods (San Diego,Calif),2014,67(3):354-363. [12] Mao X,Cai T,Olyarchuk J G. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary[J]. Bioinformatics (Oxford,England),2005,21(19):3787-3793. doi: 10.1093/bioinformatics/bti430 [13] Xing W,Qi Z,Huang C,et al. Genome-wide identification of lncRNAs and mRNAs differentially expressed in non-functioning pituitary adenoma and construction of an lncRNA-mRNA co-expression network[J]. Biology Open,2019,8(1):2046. [14] Chin C H,Chen S H,Wu H H,et al. cytoHubba:Identifying hub objects and sub-networks from complex interactome[J]. BMC Systems Biology,2014,8(4):11. [15] Ding H,Liu J,Zou R,et al. Long non-coding RNA TPTEP1 inhibits hepatocellular carcinoma progression by suppressing STAT3 phosphorylation[J]. Journal of Experimental & Clinical Cancer Research:CR,2019,38(1):189. -






 下载:
下载: