Expression and Significance of Plasma miR-1181 in Patients with Type 2 Diabetes Mellitus
-
摘要:
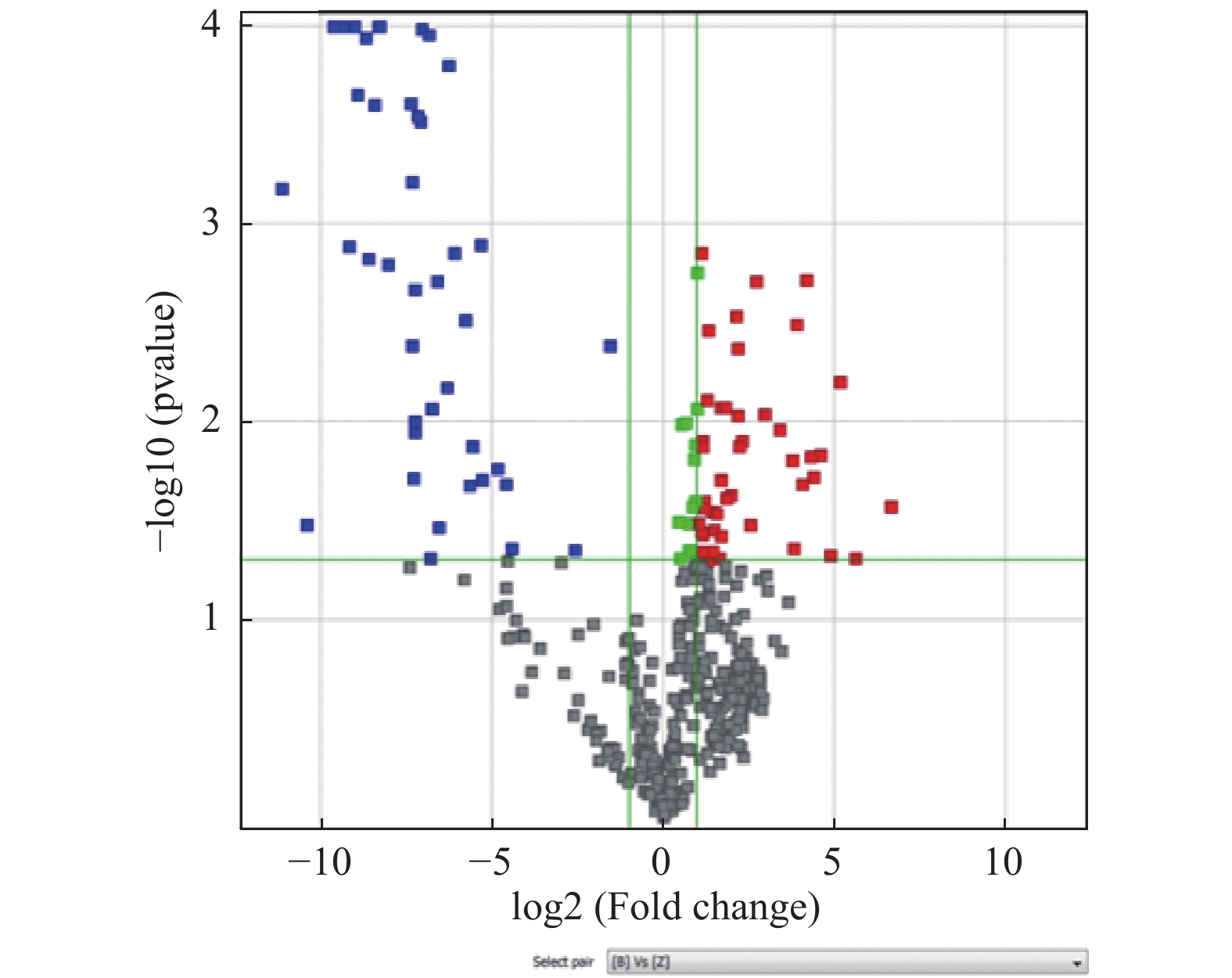
目的 miRNA可作为预测糖尿病发生和发展的生物标志物,探索miR-1181在2型糖尿病(Type 2 Diabetes Mellitus,T2DM)患者的表达情况。 方法 运用Agilent miRNA芯片对T2DM患者(n = 5)和正常对照组(n = 5)血浆样本进行差异筛选,荧光定量PCR技术(quantitative Real Time PCR,qRT-PCR)验证出差异表达的miRNA;通过生物信息学方法预测其靶基因并绘制miRNA-靶基因-代谢通路关系网络图,扩大样本检测血浆中差异表达的miRNA:miR-1181和靶基因MAP2K2、MAPK12表达水平。 结果 miRNA芯片筛选和qRT-PCR验证出与T2DM相关的差异表达miRNA:miR-1181,生物信息学分析得出miR-1181靶基因有CCND1、PI3KR2、MAP2K2和MAPK12;T2DM患者血浆中 miR-1181的表达水平明显下降(P < 0.001),而靶基因MAPK12 mRNA的表达水平明显升高(P < 0.01)。 结论 2型糖尿病患者血浆miR-1181水平降低,可能与其抑制靶基因MAPK12对MAPK通路影响有关。 Abstract:Objective To verify that miRNA can be used as biomarkers to predict the occurrence and development of diabetes mellitus (DM), and to explore the expression of miR-1181 in patients with type 2 diabetes mellitus (Type 2 Diabetes Mellitus). Methods Agilent miRNA chip was used to screen plasma samples from patients with T2DM (n = 5) and normal control group (n = 5). The differentially expressed miRNA were verified by fluorescence quantitative PCR (qRT-PCR). The target genes were predicted by bioinformatics methods, and the relationship network diagram of miRNA - target gene - metabolic pathway was drawn. The differentially expressed miRNA: miR-1181, MAP2K2 and MAPK12 in plasma were detected by expanding samples. Results miRNA chip screening and qRT-PCR verified the differentially expressed miRNA associated with T2DM: miR-1181.Bioinformatics analysis showed that the target genes of miR-1181 were CCND1, PI3KR2, MAP2K2 and MAPK12. The expression of miR-1181 in the plasma of T2DM was significantly decreased (P < 0.001). While expression of MAPK12 mRNA was increased (P < 0.01). Conclusion The decrease of plasma miR-1181 in T2DM patients may be related to inhibiting effect of target gene MAPK12 on MAPK pathway. -
Key words:
- miR-1181 /
- Type 2 Diabetes /
- Target Genes /
- MAPK12
-
表 1 qRT-PCR 引物及退火温度列表
Table 1. qRT-PCR Primer and annealing temperature list
引物名称 序列(5′to3′) 退火温度Tm(℃) U6-F CTCGCTTCGGCAGCACATATACT 58 U6 -R ACGCTTCACGAATTTGCGTGTC cel-miR-39-5p-RT GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTATTAC cel-miR-39-5p-F TGGGAGCTGATTTCGTCTTG 59.7 hsa-mir-1181-RT GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGGCTC hsa-mir-1181-F AATAACCGTCGCCGCCACCCG 72.5 GAPDH-F: GGTTGTCTCCTGCGACTTCA 58.2 GAPDH-R TGGTCCAGGGTTTCTTACTCC 58.2 MAP2K2-F: CACTATCAACCCTACCATCGC 57.3 MAP2K2-R: CGCTTCCTTTGCTGCTCAT 58.8 MAPK12-F TCTGTCCTGACCAACGCAA 57.6 MAPK12-R AAGGGACTCAAAGTATGGATGG 57.6 表 2 患者和对照组临床资料比较[(
$\bar x \pm s$ ),M(P25,P75)]Table 2. Clinical data of patients between two groups
项目 对照组(n = 65) T2DM组(n = 65) 性别(男/女) 34/31 36/29 年龄(岁) 33.5 ± 11.69 56.27 ± 12.28▲ BMI(kg/m2) 22.09 ± 3.18 23.80 ± 3.58▲ FPG(mmol/L) 5.0(4.72,5.44) 7.05(5.45,9.7) ▲ TC(mmol/L) 4.47 ± 0.52 4.21 ± 1.0▲ TG(mmol/L) 1.1(0.79,1.64) 1.61 (1.3,2.4) ▲ LDL(mmol/L) 2.48 ± 0.44 2.88 (2.41,3.11) ▲ HDL(mmol/L) 1.37(1.21,1.68) 0.98 (0.83,1.12) ▲ UA(μmol/L) 344.01(300.25,382.5) 320.3(284.4,354.15) Cr(μmol/L) 76.0(62.5,85.25) 62.75 (52.03,70.7) eGFR(ml/min*1.73m2) 106.76 ± 17.04 108.97 ± 28.15 UACR(mg/g) - 26.8(12.34,50.86) 注:正态性分布资料以(${{\bar x} } \pm s$)表示,非正态性资料以M(P25,P75)表示,与对照组比较,▲P < 0.05。BMI:体重指数;FPG:空腹血糖;TC:总胆固醇;TG:甘油三酯;LDL:低密度脂蛋白;HDL:高密度脂蛋白;UA:尿酸;Cr:肌酐;eGFR:估计肾小球滤过率;UACR:尿白蛋白肌酐比。 表 3 miR-1181靶基因预测结果
Table 3. Prediction Results of miR-1181 Target Genes
miRNA Transcript Total score Total energy Max score Max energy Strand miRNA
lengthTranscript
lengthTranscript
PostionmiR-1181 ZBTB4 150 −18.25 150 −18.25 3 21 5961 5584 miR-1181 MAP2K2 157 −25.76 157 −25.76 1 21 1759 1645 miR-1181 MAPK12 156 −15.92 143 −15.92 2 21 1778 1644 -
[1] 叶莺,刘小华,严延生. 2型糖尿病患者血浆microRNA差异表达及生物信息学分析[J]. 中华医学遗传学杂志,2021,38(6):536-540. [2] Ying S Y,Chang D C,Lin S L. The microRNA[J]. Methods MolBiol,2018,1733(10):1-25. [3] Zhang N. Protocols for the analysis of microRNA expression,biogenesis,and function in immune cells[J]. Curr Protoc Immunol,2019,126(1):e78. [4] 周建萍,毛振彪,吴信华. 微RNA-9对胃癌细胞内CDX2基因表达的影响[J]. 国际消化病杂志,2018,38(5):314-319. doi: 10.3969/j.issn.1673-534X.2018.05.009 [5] 郭青松,王鹏,黄龑,等. 微小RNA-30b对胰腺癌干细胞迁移和侵袭的调控作用[J]. 中华医学杂志,2019,99(38):3019-3023. doi: 10.3760/cma.j.issn.0376-2491.2019.38.011 [6] Hua X,Fan K C. Down-regulation of miR-1181 indicates a dismal prognosis for nasopharyngeal carcinoma and promoted cell proliferation and metastasis by modulating Wnt/beta-catenin signaling[J]. Eur Rev Med Pharmacol Sci,2019,23(3):1077-1086. [7] Ruan L. Serum miR-1181 and miR-4314 associated with ovarian cancer:MiRNA microarray data analysis for a pilot study[J]. Eur J Obstet Gynecol Reprod Biol,2018,222(1):31-38. [8] Jiang J. MiR-1181 inhibits stem cell-like phenotypes and suppresses SOX2 and STAT3 in human pancreatic cancer[J]. Cancer Lett,2015,356(2PtB):962-70. [9] Wang J. miR-1181 inhibits invasion and proliferation via STAT3 in pancreatic cancer[J]. World J Gastroenterol,2017,23(9):1594-1601. doi: 10.3748/wjg.v23.i9.1594 [10] Nam E J. MicroRNA profiling of a CD133(+) spheroid-forming subpopulation of the OVCAR3 human ovarian cancer cell line[J]. BMC Med Genomics,2012,5(18):1-9. [11] Ruan L. Serum miR-1181 and miR-4314 associated with ovarian cancer:MiRNA microarray data analysis for a pilot study[J]. Eur J Obstet Gynecol Reprod Biol,2018,222(10):31-38. [12] 王杰. miRNA-1181通过抑制STAT3影响人胰腺癌细胞的侵袭转移能力[J]. 贵州医科大学学报,2016,41(12):1387-1392. [13] Van Allen EM,Wagle N,Sucker A,et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma[J]. Cancer Discov,2014,4(1):94-109. doi: 10.1158/2159-8290.CD-13-0617 [14] Zhong W,Zhu H,Sheng F,et al. Activation of the MAPK11/12/13/14 (p38 MAPK) pathway regulates the transcription of autophagy genes in response to oxidative stress induced by a novel copper complex in HeLa cells[J]. Autophagy,2014,10(7):1285-1300. doi: 10.4161/auto.28789 [15] 窦艳,邱鹏,王丽. NEK2及ERK2在不同病理类型乳腺癌中的表达及意义[J]. 临床合理用药杂志,2019,12(14):120-121. -






 下载:
下载: