The Genetic Characteristic Analysis of the VP1 Gene of Coxsackievirus A6 in Qujing Prefecture of Yunnan Province in 2020
-
摘要:
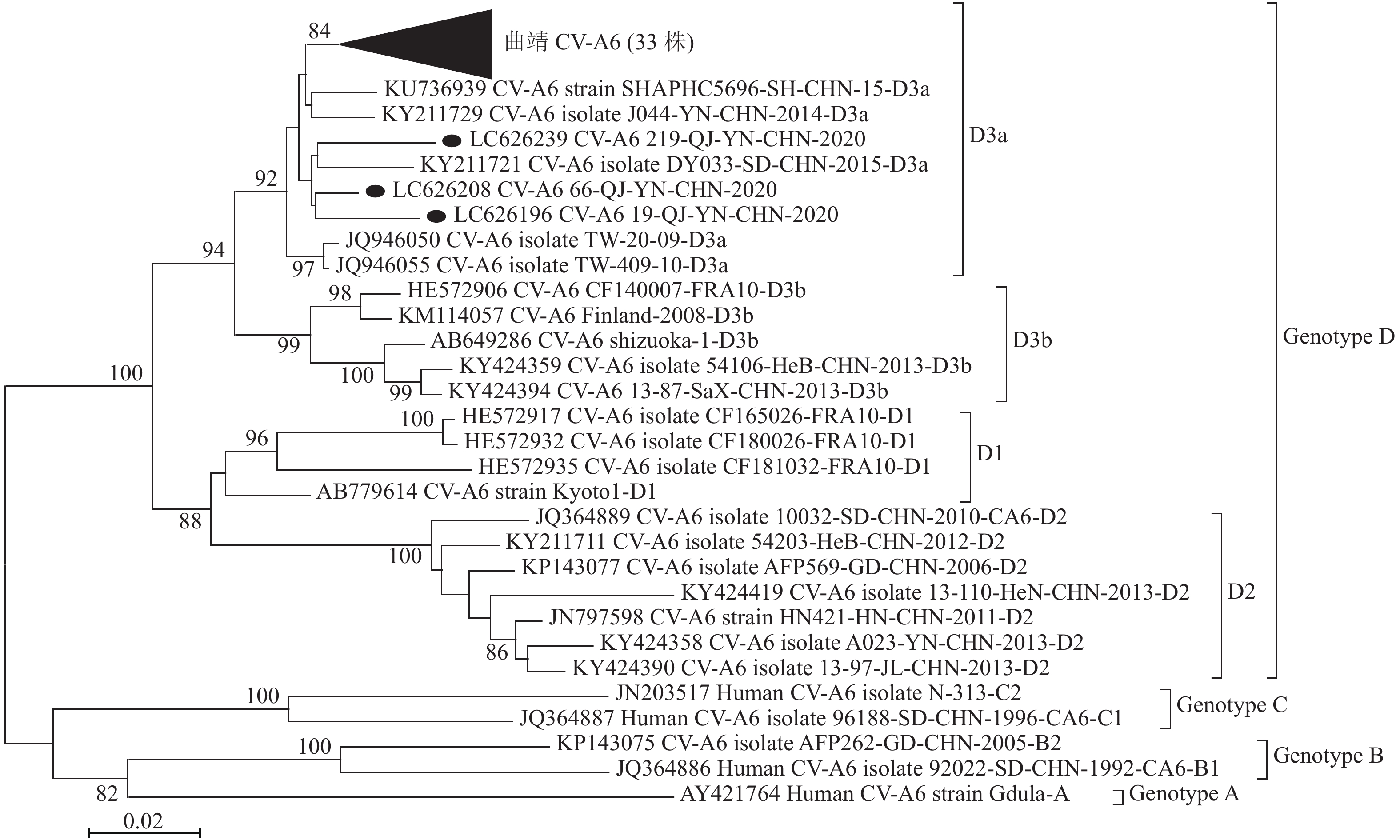
目的 对2020年云南省曲靖地区手足口病(hand,foot and mouth disease, HFMD)病例中的柯萨奇病毒A组6型(Coxsackievirus A6, CV-A6)VP1区基因特征进行分析。 方法 (1)直接从HFMD病例粪便标本中提取人类肠道病毒(human enteroviruses,EVs)RNA,先用MD91/OL68-1引物对病毒VP4/VP2结合区基因进行逆转录-聚合酶链反应(Reverse-Transcription Polymerase Chian Reaction, RT-PCR)和测序,确定病毒型别,再用各型病毒的相关引物扩增VP1区全序并测序,按测序定型标准对EVs进行VP1区定型;(2)按文献下载基因库中CV-A6病毒VP1区基因参考序列,用MEGA 5.2软件构建系统进化树,进行基因特征和分子流行病学分析。 结果 2020年共从曲靖地区HFMD 病例500份标本中检测到49株EVs,EVs阳性率为9.80%(49/500)。49株EVs中,A组病毒(enterovirus species A,EV-A) 48份,占阳性总数的97.96%(48/49),B组病毒(enterovirus species B,EV-B)1份,占2.04%(1/49)。A组病毒共4个血清型,其中CV-A4 3株,占6.12%(3/49);CV-A6 36株,占73.47%(36/49);CV-A10 5株,占10.20%(5/49);CV-A16 4株,占8.16%(4/49)。B组病毒只检测到1株,一个血清型(CV-B4)。用MEGA 5.2软件对36株CV-A6病毒的VP1基因完整序列(915bp)和Yang Song、Mizuta K等发表的26株参考株序列共62株病毒作基因进化分析,曲靖地区36株均为D3a亚型。 结论 2020年云南省曲靖地区HFMD流行以CV-A6(占73.47%)为主。基因进化分析表明,跟以往中国内地的情况一样,云南省曲靖地区2020年流行的CV-A6为D3a基因亚型,加强CV-A6病毒的实验室和分子流行病学监测是今后曲靖地区乃至云南省HFMD监测的重要工作。 Abstract:Objective To analyze the genetic characteristics of the VP1 gene of coxsackievirus A6 (CV-A6) from hand, foot and mouth disease (HFMD) cases collected from Qujing prefecture in 2020. Methods (1) Virus RNA was directly abstracted from stool samples of HFMD cases and virus VP4/VP2 junction region sequence was firstly amplified and sequenced by MD91 and OL68-1 primer pairs, then the virus serotype was determined. The virus entire VP1 gene sequences were determined by relative primer pairs according to the references. (2) The reference sequences of CV-A6 virus entire VP1 gene were downloaded from the GenBank and the phylogenetic tree was constructed and the genetic characteristics and molecular epidemiology were analyzed. Results In 2020, a total of 49 strains of enteroviruses (EVs) were identified with a positive rate of 9.80% (49/500). Among 49 EVs strains, 48 were enterovirus species A (EV-A), accounting for 97.96% (48/49), 1 was enterovirus species B (EV-B), accounting for 2.04% (1/49). A total of 4 serotypes were identified from EV-A, among which, 3 were CV-A4, accounting for 6.12% (3/49); 36 were CV-A6, accounting for 73.47% (36/49); 5 were CV-A10, accounting for 10.20% (5/49) and 4 were CV-A16, accounting for 8.16% (4/49). Only one strain of EV-B viruses was identified (coxsackievirus B4, CV-B4), accounting for 2.04% (1/49). The phylogenetic characteristics of the entire VP1 gene sequences of 36 strains of CV-A6 identified in this study and 26 reference strains published by Yang Song and Mizuta K were analyzed by MEGA software 5.2. The results showed that all 36 Qujing strains belonged to D3a sub-genotype. Conclusion The outbreaks of HFMD in Qujing prefecture in 2020 are mainly caused by CV-A6, accounting for 73.47%. Phylogenetic analysis shows that, similar to the situation in Mainland of China during the past years, the sub-genotype D3a of CV-A6 is the predominant virus in this outbreak. Strengthening the laboratory and molecular epidemiological surveillance is the important work in Qujing prefecture and even in the whole Yunnan province in the future. -
Key words:
- Hand foot and mouth disease (HFMD) /
- Pathogen /
- Coxsackievirus A6 /
- VP1 gene /
- Genetic analysis
-
表 1 云南省曲靖地区2020年500份标本中HFMD病原分离率(%)
Table 1. Etiological isolation rates from 500 samples of HFMD patients,Qujing prefecture,Yunnan province,2020 (%)
病毒组别 病毒型别 分离数 分离率 EV-A CV-A4 3 6.12 CV-A6 36 73.47 CV-A10 5 10.2 CV-A16 4 8.16 EV-B CV-B4 1 2.04 表 2 2019~2020年曲靖地区HFMD病原检测比较n(%)
Table 2. Pathogen detection from HFMD patients,Qujing prefecture,2019~2020 n(%)
病毒组别 病毒型别 2019年 2020年 EV-A CV-A4 1(1.61) 3(6.12) CV-A6 8(12.90) 36(73.47) CV-A10 21(33.87) 5(10.20) CV-A16 12(19.35) 4(8.16) EV-B CV-B4 − 1(2.04) E11 3(4.84) − E12 3(4.84) − E14 1(1.61) − E30 2(3.22) − EV-C PV1 11(17.74) − 注:“−”表示无。 -
[1] 国家卫生健康委. 手足口病诊疗指南(2018年版)[J]. 中国病毒病杂志,2018,8(5):347-352. [2] Yong Zhang,Xiao-Juan Tan,Hai-Yan Wang,et al. An outbreak of hand,foot,and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong,China[J]. J Clin Virol,2009,44(4):262-267. doi: 10.1016/j.jcv.2009.02.002 [3] Yang F,Ren L,Xiong Z,et al. Enterovirus 71 outbreak in the people’s republic of China in 2008[J]. J Clin Microbiol,2009,47(7):2351-2352. doi: 10.1128/JCM.00563-09 [4] Li L,He Y,Yang H,et al. Genetic characteristics of human enterovirus 71 and coxsackievirus A16 circulating from 1999 to 2004 in shenzhen,people’s republic of China[J]. J Clin Microbiol,2005,43(8):3835-3839. doi: 10.1128/JCM.43.8.3835-3839.2005 [5] Liu W,Wu S,Xiong Y,et al. Co-circulation and genomic recombination of coxsackievirus A16 and enterovirus 71 during a large outbreak of hand,foot and mouth disease in central China[J]. PLoS One,2014,9(4):96051. doi: 10.1371/journal.pone.0096051 [6] Song Y,Zhang Y,Ji T,et al. Persistent circulation of coxsackievirus A6 of genotype D3 in mainland of China between 2008 and 2015[J]. Sci Rep,2017,7(1):5491. doi: 10.1038/s41598-017-05618-0 [7] 陈炜,翁育伟,何文祥,等. 2011~2013年福建省手足口病相关病原柯萨奇病毒A组6型的分子流行病学研究[J]. 病毒学报,2014,30(6):624-629.Chen Wei,Weng Yu-wei,He Wen-xiang,et al. Molecular epidemiology of HFMD-associated pathogen coxsackievirus A6 in Fujian province,2011-2013[J]. Chin J Virol,2014,30(6):624-629. [8] Ishiko H,Shimada Y,Yonaha M,et al. Molecular diagnosis of human enteroviruses by phylogeny-based classification by use of the VP4 sequence[J]. J Infect Dis,2002,185(6):744-754. doi: 10.1086/339298 [9] 李桂满,罗春蕊,刘艳艳,等. 基于VP4/VP2结合区基因序列对手足口病病原分型的初步研究[J]. 中国病毒病杂志,2021,11(4):271-274.Li Guiman,Luochun,Liu Yanyan,et al. Preliminary study on etiological typing of hand,foot and mouth disease based on VP4/VP2 junction region sequences[J]. Chin J Viral Dis,2021,11(4):271-274. [10] Oberste M S,Maher K,Williams A J,et al. Species-specific RT-PCR amplification of human enteroviruses:A tool for rapid species identification of uncharacterized enteroviruses[J]. J Gen Virol,2006,87(1):119-128. doi: 10.1099/vir.0.81179-0 [11] Obester M S,K Maher,D R Kilpatrick,et al. Typing of human enteroviruses by partial sequencing of VPl[J]. J Clin Microbiol,1999,37(5):1288-1293. doi: 10.1128/JCM.37.5.1288-1293.1999 [12] Obester M S,K Maher,D R Kilpatrick,et al. Molecular evolution of the human enteroviruses:Correlation of serotype with VP1 sequence and application to picornavirus classification[J]. J Virol,1999,73(3):1941-1948. doi: 10.1128/JVI.73.3.1941-1948.1999 [13] Mizuta K,Tanaka S,Komabayashi K,et al. Phylogenetic and antigenic analyses of coxsackievirus A6 isolates in Yamagata,Japan between 2001 and 2017[J]. Vaccine,2019,37(8):1109-1117. doi: 10.1016/j.vaccine.2018.12.065 [14] 李桂满,张勇,刘艳艳,等. 云南省曲靖地区手足口病病原学监测(2019年)[J]. 国际流行病学传染病学杂志,2021,48(5):362-367. doi: 10.3760/cma.j.cn331340-20210208-00027 [15] Gilbert Dalldorf,Grace M Sickles,Hildegard Plager,et al. A virus recovered from the feces of poliomyelitis patients pathogenic for suckling mice[J]. J Exp Med,1949,89(6):567-582. doi: 10.1084/jem.89.6.567 [16] Menghua Xu,Lijun Su,Lingfeng Cao,et al. Genotypes of the enterovirus causing hand foot and mouth disease in Shanghai,China,2012-2013[J]. Plos One,2015,10(9):e0138514. doi: 10.1371/journal.pone.0138514 -






 下载:
下载: