Population Genetic Analysis of 18 Autosomal STR Loci in Minjia Branch of Bai Nationality in Yunnan Province, Southwest China
-
摘要:
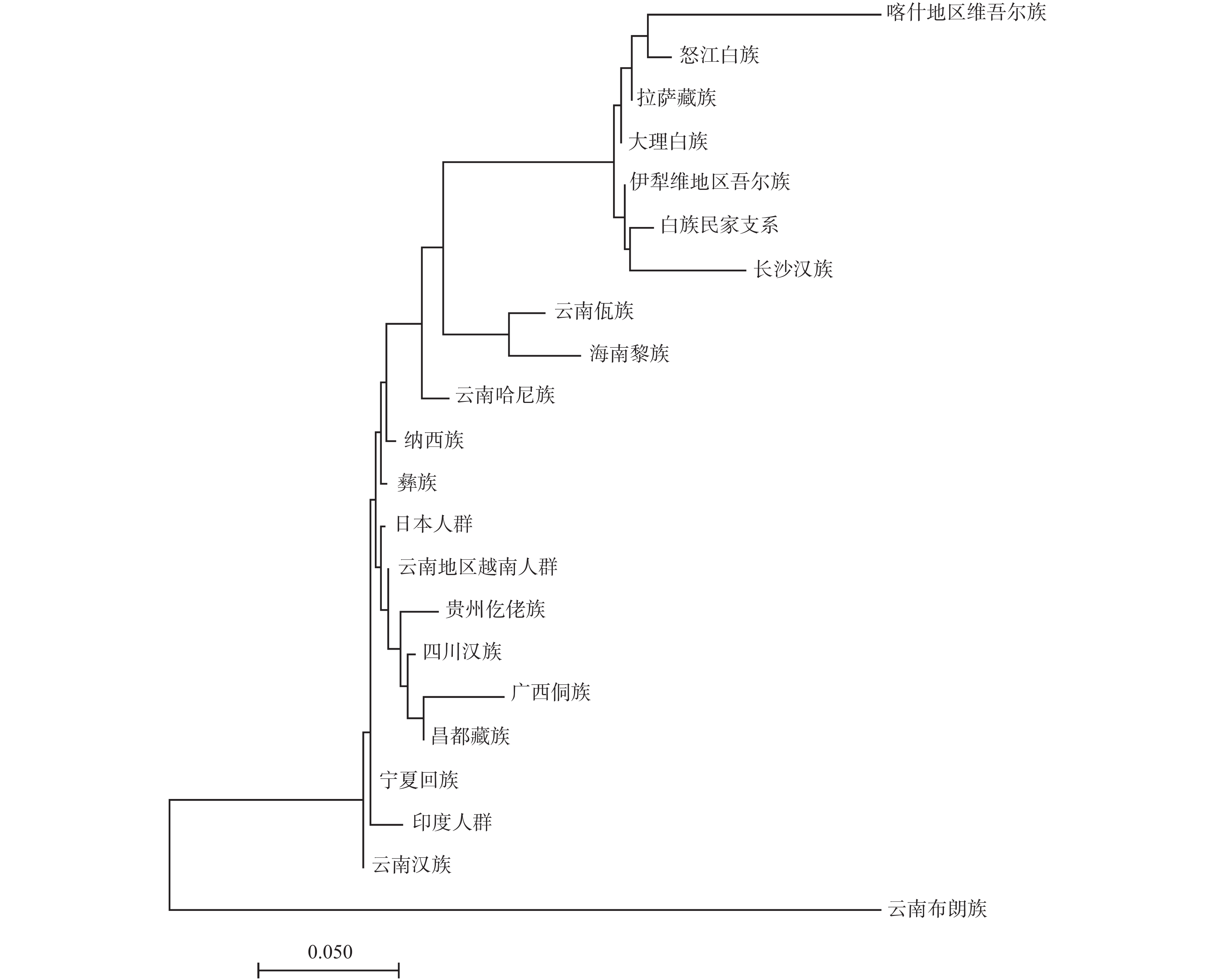
目的 调查云南白族民家支系2 323个健康无亲缘关系个体18个STR基因座的遗传多态性,计算群体遗传学参数,建立云南白族民家支系群体的遗传学基础数据,为司法鉴定工作中的亲子鉴定和个体识别提供可靠的科学依据。 方法 收集2323份云南白族民家支系人群无关个体样本,采用DNATyperTM19试剂盒,进行直接PCR复合扩增,用AB 3130XL自动遗传分析仪进行毛细管电泳分离,用GeneMapper ID-X 1.5软件对分离的STR进行分型。使用Modified-Powerstats软件统计等位基因频率,进行Hardy-Weinberg平衡(HWE)检验,计算匹配概率等法医学参数。使用Arlequin软件计算Fst和P值。使用Phylip软件计算Nei’s遗传距离。使用SPSS软件进行多维尺度(MDS)分析。应用Mega软件构建相邻连接(NJ)系统发育树。 结果 18个STR基因座中共观察到230个等位基因和1073种基因型。除D13S317基因座不符合Hardy-Weinberg平衡(P = 0.0008)外,其余基因座等位基因频率和基因型频率均符合Hardy-Weinberg平衡(P>0.05/18 = 0.0028)。18个STR基因座的累积个人识别能力(CDP)达0.999999999,累积非父排除概率(CPE)达0.99999994055。Fst值、Nei's遗传距离以及NJ系统发育树的结果均提示,白族民家支系与云南布朗族和云南佤族相距较远,而与大理白族和怒江白族相距较近。 结论 上述18个STR基因座在云南白族民家支系群体中具有高度多态性,可用于司法鉴定和群体遗传结构研究。 Abstract:Objective To investigate the genetic polymorphism of 18 autosomal short tandem repeats (STR)loci in 2323 healthy unrelated individuals of Minjia branch of Bai nationality in Yunnan, calculate the population genetic parameters, and establish the genetic data of Minjia branch of Bai nationality in Yunnan, so as to provide the scientific basis for the parental identification and individual identification of forensic material evidence. Methods A total of 2323 unrelated individuals from Minjia branch of Bai nationality population were collected. DNA TyperTM 19 kit was used for direct PCR compound amplification. AB 3130XL automatic genetic analyzer was used to separate the PCR compound amplification products by capillary electrophoresis. STR typing was performed with GeneMapper ID-x 1.5 software. Forensic parameters, such as allele frequency and matching probability, were calculated by Modified Powerstats software, and the hardy-Weinberg equilibrium (HWE) test was performed. The Fst and P values were calculated using Arlequin software. Nei’s genetic distance was calculated using Phylip software. Multi-dimensional scale (MDS) analysis was performed using SPSS software. The Neighbor-Joining (NJ) phylogenetic tree was constructed by Mega software. Results A total of 230 alleles and 1073 genotypes were observed at 18 STR loci. Except that D13S317 loci was not in accord with Hardy-Weinberg equilibrium (P = 0.0008), the allele frequencies and genotype frequencies of other loci were consistent with Hardy-Weinberg equilibrium (P > 0.05/20 = 0.0025). The cumulative discrimination power (CDP) and cumulative probability of exclusion (CPE) of 18 STR loci were 0.999999999 and 0.99999994055, respectively. The results of Fst, Nei’s genetic distance and NJ phylogenetic tree indicated that the Minjia branch of Bai nationality was close to the Bai nationality in Nujiang and the Bai nationality in Dali, but far away from the Bulang nationality in Yunnan and Va nationality in Yunnan. Conclusion The 18 STR loci mentioned above show the high genetic polymorphism in the Minjia branch of Bai nationality and they can be used for the forensic identification and population genetics research. -
表 1 白族民家支系18个STR基因座的等位基因频率及法医学参数(n = 2323)(1)
Table 1. Allele frequencies and forensic parameters of 18 STRs in Minjia branch of Bai ethnic minority (n = 2323)(1)
Allele D3S1358 D6S1043 D13S317 Penta E D16S539 D18S51 D2S1338 CSF1PO TH01 vWA D21S11 D7S820 D5S818 TPOX D8S1179 D12S391 D19S433 FGA 5 0.0458 0.0009 6 0.0002 0.0846 7 0.0022 0.0011 0.0019 0.2833 0.0013 0.0217 0.0009 7.3 8 0.0011 0.2749 0.0110 0.0075 0.0009 0.0628 0.1291 0.0006 0.5093 8.1 9 0.0028 0.1369 0.0065 0.2400 0.0002 0.0426 0.4957 0.0719 0.0773 0.1244 0.0004 0.0037 9.1 0.0006 9.3 0.0454 10 0.0353 0.1440 0.0347 0.1104 0.0009 0.2374 0.0271 0.1614 0.1920 0.0239 0.0945 0.0006 10.1 0.0015 11 0.1197 0.2322 0.1894 0.3037 0.0026 0.2443 0.0002 0.3259 0.3250 0.3117 0.0771 0.0028 12 0.0004 0.1438 0.1627 0.1257 0.2163 0.0224 0.3898 0.2658 0.2331 0.0295 0.1156 0.0357 12.2 0.0140 13 0.0013 0.1382 0.0366 0.0560 0.1089 0.2086 0.0766 0.0002 0.0362 0.1332 0.0004 0.2548 0.2785 13.2 0.0517 14 0.0418 0.1203 0.0105 0.0859 0.0123 0.2023 0.0058 0.2286 0.0058 0.0157 0.1901 0.2499 14.2 0.0977 15 0.3549 0.0129 0.0803 0.0009 0.1950 0.0006 0.0276 0.0004 0.0011 0.1722 0.0118 0.0687 15.2 0.1429 16 0.3487 0.0009 0.0986 0.1319 0.0149 0.1746 0.0002 0.0755 0.0065 0.0133 16.2 0.0355 17 0.1754 0.0502 0.0605 0.0809 0.0493 0.2566 0.0161 0.1009 0.0017 0.0006 17.2 0.0024 17.3 0.0004 18 0.0710 0.1651 0.0678 0.0402 0.1009 0.2058 0.0037 0.2660 0.0006 0.0327 18.2 0.0002 0.0002 18.3 0.0009 19 0.0058 0.1455 0.0489 0.0390 0.1754 0.0876 0.2004 0.0439 20 0.0006 0.0478 0.0433 0.0224 0.1274 0.0181 0.1573 0.0654 20.2 0.0002 0.0006 20.3 0.0022 21 0.0062 0.0245 0.0183 0.0512 0.0011 0.0994 0.1009 21.1 0.0002 21.2 0.0004 0.0077 21.3 0.0071 注:Pm: 随机匹配概率; DP: 个体识别能力; PIC: 多态性信息含量; PE: 非父排除概率;TPI: 典型亲权指数; Ho: 观测值杂合度;p-value: Hardy-Weinberg平衡(HWE)检验P值. 表 1 白族民家支系18个STR基因座的等位基因频率及法医学参数(n = 2323)(2)
Table 1. Allele frequencies and forensic parameters of 18 STRs in Minjia branch of Bai ethnic minority (n = 2323)(2)
Allele D3S1358 D6S1043 D13S317 Penta E D16S539 D18S51 D2S1338 CSF1PO TH01 vWA D21S11 D7S820 D5S818 TPOX D8S1179 D12S391 D19S433 FGA 22 0.0004 0.0084 0.0187 0.0420 0.0880 0.1707 22.2 0.0065 23 0.0043 0.0084 0.2260 0.0536 0.2273 23.2 0.0123 24 0.0045 0.0054 0.1446 0.0075 0.1672 24.2 0.0170 25 0.0019 0.0022 0.0570 0.0060 0.0895 25.2 0.0024 26 0.0009 0.0006 0.0095 0.0009 0.0426 26.2 0.0013 27 0.0015 0.0024 0.0065 28 0.0002 0.0508 0.0045 28.2 0.0166 29 0.2484 29.2 0.0028 30 0.2652 30.1 0.0002 30.2 0.0230 30.3 0.0006 31 0.1007 31.2 0.0687 32 0.0284 32.2 0.1337 33 0.0024 33.2 0.0463 34.2 0.0093 35.2 0.0006 Pm 0.1292 0.0283 0.0654 0.0166 0.0837 0.0408 0.0348 0.1210 0.1654 0.0691 0.0495 0.0854 0.0823 0.2023 0.0468 0.0476 0.0542 0.0334 DP 0.8708 0.9717 0.9346 0.9834 0.9163 0.9592 0.9652 0.8790 0.8346 0.9309 0.9505 0.9146 0.9177 0.7977 0.9532 0.9524 0.9458 0.9666 PIC 0.6652 0.8639 0.7744 0.8977 0.7446 0.8311 0.8453 0.6781 0.6115 0.7703 0.8087 0.7400 0.7456 0.5641 0.8146 0.8131 0.7976 0.8491 PE 0.4315 0.7312 0.5548 0.7939 0.5403 0.6708 0.7029 0.4478 0.3745 0.5844 0.6384 0.5679 0.5610 0.3182 0.6401 0.6393 0.6148 0.7191 TPI 1.6785 3.7958 2.2294 4.9637 2.1509 3.0809 3.4263 1.7388 1.4872 2.4048 2.7921 2.3046 2.2641 1.3229 2.8056 2.7988 2.6101 3.6297 Ho 0.7021 0.8683 0.7757 0.8993 0.7675 0.8377 0.8541 0.7124 0.6638 0.7920 0.8209 0.7830 0.7792 0.6220 0.8218 0.8214 0.8084 0.8622 HWE(p-value) 0.1687 0.2066 0.0008 0.3332 0.1724 0.1348 0.3628 0.1954 0.7213 0.3084 0.3011 0.3009 0.9749 0.6396 0.0800 0.1155 0.1521 0.8597 注:Pm:随机匹配概率;DP:个体识别能力;PIC:多态性信息含量;PE:非父排除概率;TPI:典型亲权指数;Ho:观测值杂合度;p-value:Hardy-Weinberg平衡(HWE)检验P值。 表 2 白族民家支系与21个已发表人群的Fst和P值(1)
Table 2. Pairwise Fst and P values between Minjia branch of Baiethnic minority and 21 published populations (1)
民族 STR基因座 CSF1PO D2S1338 D3S1358 D5S818 D7S820 Fst P Fst P Fst P Fst P Fst P 喀什地区维吾尔族 0.00323 < 0.00001# 0.00541 < 0.00001# 0.00311 < 0.00001# 0.00659 < 0.00001# 0.01279 < 0.00001# 云南佤族 0.00248 0.00195# 0.00708 < 0.00001# 0.01609 < 0.00001# 0.01171 < 0.00001# 0.00165 0.00684* 广西侗族 0.00068 0.04102* 0.00309 < 0.00001# 0.00422 < 0.00001# 0.00177 < 0.00001# 0.00702 < 0.00001# 海南黎族 0.00050 0.19922 0.00632 < 0.00001# 0.00509 < 0.00001# 0.00268 0.00488* 0.00223 0.01660* 昌都藏族 0.00117 0.00879* 0.00029 0.09473 0.28350 < 0.00001# 0.00140 0.00488* 0.00051 0.06543 四川汉族 −0.00129 0.83691 −0.00031 0.55566 −0.00077 0.60059 −0.00130 0.89355 −0.00030 0.46680 云南布朗族 0.00449 0.04102* 0.00557 0.00391* 0.00945 0.00293# 0.00376 0.04688* −0.00037 0.48047 云南哈尼族 0.00217 0.02539* 0.00441 < 0.00001# 0.00162 0.04883* 0.00318 0.00195# 0.00168 0.02148* 云南地区越南人群 0.01493 < 0.00001# 0.00689 < 0.00001# 0.02162 < 0.00001# 0.00130 0.07617 0.02601 < 0.00001# 贵州仡佬族 −0.00062 0.63477 −0.00046 0.66992 0.00161 0.09082 −0.00056 0.66504 0.00238 0.04004* 云南汉族 −0.00025 0.72852 0.00050 0.04395* 0.00083 0.03711* 0.00056 0.07910 0.00369 < 0.00001# 彝族 0.00007 0.33105 0.00031 0.26758 0.00074 0.19531 0.00050 0.25000 −0.00095 0.89062 纳西族 −0.00095 0.90430 −0.00023 0.57617 −0.00006 0.36816 0.00000 0.40332 0.00037 0.24023 日本人群 0.00214 0.01562* 0.01237 < 0.00001# 0.00326 0.00977* 0.00084 0.10352 0.00425 < 0.00001# 印度人群 0.00364 0.02734* 0.00318 0.00488* 0.00547 0.00488* 0.00498 0.00391* 0.01890 < 0.00001# 宁夏回族 −0.00041 0.64453 0.00052 0.12402 0.00117 0.06641 0.00042 0.19043 0.00183 0.01562* 伊犁维吾尔族 0.00387 0.03223* 0.10822 < 0.00001# 0.00544 0.01367* 0.00796 0.00195# 0.00851 < 0.00001# 长沙汉族 0.00010 0.27051 0.00039 0.07031 0.00288 < 0.00001# 0.00032 0.13379 0.00124 0.01074* 拉萨藏族 −0.00197 0.85840 0.00085 0.23828 0.00083 0.25391 0.00214 0.13379 0.00019 0.36230 怒江白族 0.00440 0.09473 −0.00277 0.93262 −0.00212 0.71289 0.00262 0.17969 −0.00293 0.86816 大理白族 −0.00020 0.52148 −0.00050 0.97852 0.00034 0.20312 0.00006 0.35547 −0.00043 0.85742 即使在Bonforroni校正后,本研究人群与比较人群之间的Fst值也存在显著差异(#P < 0.05/15 = 0.0033);*P < 0.05。 表 2 白族民家支系与21个已发表人群的Fst和P值(2)
Table 2. Pairwise Fst and P values between Minjia branch of Bai ethnic minority and 21 published populations (2)
民族 STR基因座 D8S1179 D13S317 D16S539 D18S51 D19S433 Fst P Fst P Fst P Fst P Fst P 喀什地区维吾尔族 0.00074 0.00781* 0.01187 < 0.00001# 0.00128 0.00195# 0.00397 < 0.00001# 0.00308 < 0.00001# 云南佤族 0.00380 < 0.00001# 0.00854 < 0.00001# 0.00645 < 0.00001# 0.00626 < 0.00001# 0.00365 < 0.00001# 广西侗族 0.01678 < 0.00001# 0.00722 < 0.00001# 0.00182 < 0.00001# 0.00464 < 0.00001# 0.00050 0.04395* 海南黎族 0.01014 < 0.00001# 0.00329 0.00293# 0.00163 0.03223* 0.00464 < 0.00001# 0.00048 0.17383 昌都藏族 0.00132 0.00098# 0.13054 < 0.00001# 0.00113 0.00879* 0.00509 < 0.00001# 0.00144 < 0.00001# 四川汉族 −0.00053 0.58496 −0.00084 0.73633 0.00581 0.00293# 0.00041 0.24707 −0.00102 0.83691 云南布朗族 0.00219 0.09961 0.00074 0.27734 0.00351 0.07031 −0.00175 0.93945 0.00425 0.01953* 云南哈尼族 0.00493 < 0.00001# 0.00244 0.00391* 0.00457 0.00098# 0.00544 < 0.00001# 0.00236 0.00293# 云南地区越南人群 0.03193 < 0.00001# 0.03207 < 0.00001# 0.01673 < 0.00001# 0.00364 0.00098# 0.01629 < 0.00001# 贵州仡佬族 0.00044 0.24512 0.00053 0.21973 −0.00102 0.88086 −0.00025 0.53125 0.00009 0.37012 云南汉族 0.00442 < 0.00001# 0.00097 0.01172* 0.00094 0.01172* 0.00046 0.06152 0.00033 0.12012 彝族 0.00046 0.23145 0.00184 0.04883* −0.00035 0.55273 0.00115 0.09668 0.00155 0.05566 纳西族 0.00516 < 0.00001# −0.00113 0.99707 0.00114 0.10840 −0.00030 0.61719 −0.00018 0.51855 日本人群 0.00392 < 0.00001# 0.00143 0.03516* 0.02029 < 0.00001# 0.00092 0.05664 0.00175 0.01855* 印度人群 0.07056 < 0.00001# 0.01050 < 0.00001# 0.00631 < 0.00001# 0.14905 < 0.00001# 0.00632 < 0.00001# 宁夏回族 0.00001 0.39844 0.00136 0.04785* 0.00021 0.27539 0.00090 0.06348 −0.00029 0.65137 伊犁维吾尔族 0.00284 0.02734* 0.01284 < 0.00001# 0.00132 0.12793 0.00412 0.00391* 0.00345 0.02734* 长沙汉族 0.00194 < 0.00001# 0.00084 0.03027* 0.00143 0.00391* 0.00050 0.04688* −0.00001 0.39648 拉萨藏族 0.00047 0.28320 0.00874 0.00488* 0.00253 0.10156 0.00456 0.01953* 0.00389 0.03223* 怒江白族 −0.00176 0.73047 0.00081 0.29980 0.00307 0.14160 0.00013 0.39648 0.00289 0.13477 大理白族 0.00092 0.04004* −0.00044 0.87500 0.00015 0.29590 −0.00036 0.86328 0.00012 0.29785 注:黑色加粗的值表明,即使在Bonforroni校正后,本研究人群与比较人群之间的Fst值也存在显著差异(#P < 0.05/15 = 0.0033);*P < 0.05。 表 2 白族民家支系与21个已发表人群的Fst和P值(3)
Table 2. Pairwise Fst and P values between Minjia branch of Bai ethnic minority and 21 published populations (3)
民族 STR基因座 D21S11 FGA TH01 TPOX vWA Fst p Fst P Fst P Fst p Fst P 喀什地区维吾尔族 0.00410 < 0.00001# 0.00212 < 0.00001# 0.04589 < 0.00001# 0.00207 0.00098# 0.00704 < 0.00001# 云南佤族 0.00520 < 0.00001# 0.00407 < 0.00001# 0.00953 < 0.00001# 0.02182 < 0.00001# 0.19039 < 0.00001# 广西侗族 0.00069 0.00977* 0.00471 < 0.00001# 0.00171 < 0.00001# 0.00096 0.03809* 0.01037 < 0.00001# 海南黎族 0.00127 0.04297* 0.00395 < 0.00001# 0.00453 0.00098# 0.00347 0.01172* 0.00447 < 0.00001# 昌都藏族 0.00052 0.05762 0.00213 < 0.00001# 0.00136 0.00781* 0.01058 < 0.00001# 0.00314 < 0.00001# 四川汉族 0.00036 0.29492 0.00112 0.13867 −0.00142 0.92090 −0.00119 0.75684 −0.00105 0.81738 云南布朗族 0.00462 0.01074* 0.00144 0.13281 0.01066 0.00488* 0.02201 0.00098# 0.00084 0.23535 云南哈尼族 0.00526 < 0.00001# 0.00487 < 0.00001# 0.00075 0.14160 0.01292 < 0.00001# 0.00207 0.01465* 云南地区越南人群 0.00664 0.00098# 0.01939 < 0.00001# 0.00954 < 0.00001# 0.04698 < 0.00001# 0.01099 < 0.00001# 贵州仡佬族 0.00035 0.26562 −0.00033 0.58984 −0.00027 0.44922 −0.00092 0.71582 0.00086 0.17773 云南汉族 −0.00011 0.58691 0.00062 0.02441* 0.00015 0.22559 0.00046 0.12402 0.00161 0.00195# 彝族 −0.00034 0.60840 −0.00115 0.99414 0.00104 0.13965 −0.00059 0.57422 0.00060 0.21973 纳西族 0.00150 0.04590* 0.00048 0.19922 0.00039 0.25195 0.00037 0.24023 −0.00049 0.70020 日本人群 0.00346 < 0.00001# 0.00073 0.08887 0.02701 < 0.00001# 0.00314 0.01758* −0.00049 0.75488 印度人群 0.01773 < 0.00001# 0.00474 0.00098# 0.07503 < 0.00001# 0.03121 < 0.00001# 0.01042 < 0.00001# 宁夏回族 0.00024 0.26074 0.00028 0.22363 0.00024 0.22852 0.00034 0.20703 0.00080 0.09277 伊犁维吾尔族 0.06856 < 0.00001# 0.00305 0.01660* 0.05871 < 0.00001# 0.00044 0.27148 0.00944 < 0.00001# 长沙汉族 0.00015 0.22363 0.00025 0.15430 0.00040 0.12793 −0.00013 0.47656 0.00077 0.02734* 拉萨藏族 0.00042 0.32520 0.00294 0.04883* −0.00087 0.54883 0.00713 0.03125* −0.00062 0.51953 怒江白族 −0.00227 0.83887 0.00278 0.11523 −0.00052 0.42871 0.00201 0.20020 −0.00349 0.96777 大理白族 0.00150 0.00977* −0.00018 0.63574 0.00062 0.12695 −0.00016 0.47559 −0.00060 0.99121 即使在Bonforroni校正后,本研究人群与比较人群之间的Fst值也存在显著差异(#P < 0.05/15 = 0.0033);*P < 0.05。 表 3 白族民家支系与21个已发表人群的Nei's遗传距离
Table 3. Nei's standard genetic distances between Minjia branch of Bai nationality and 21 published populations.
[1] [2] [3] [4] [5] [6] [7] [8] [9] [10] [11] [12] [13] [14] [15] [16] [17] [18] [19] [20] [21] [22] 白族民家支系[1] 0.000000 喀什地区维吾尔族[2] 0.107646 0.000000 云南佤族[3] 0.114762 0.177184 0.000000 广西侗族[4] 0.145404 0.217277 0.096202 0.000000 海南黎族[5] 0.124407 0.227674 0.038474 0.096235 0.000000 昌都藏族[6] 0.118570 0.177630 0.073168 0.024859 0.085629 0.000000 四川汉族[7] 0.116581 0.180529 0.070624 0.035347 0.087920 0.005196 0.000000 云南布朗族[8] 0.415857 0.579774 0.398897 0.387094 0.397545 0.348782 0.355437 0.000000 云南哈尼族[9] 0.103179 0.203859 0.048691 0.059480 0.043724 0.034243 0.028206 0.365319 0.000000 云南地区越南人群[10] 0.098524 0.172589 0.059345 0.032926 0.076531 0.007025 0.008501 0.329172 0.024481 0.000000 贵州仡佬族[11] 0.115547 0.149589 0.067855 0.049599 0.093779 0.016923 0.018700 0.360752 0.049616 0.016962 0.000000 云南汉族[12] 0.097612 0.181988 0.058948 0.043558 0.072595 0.014636 0.014362 0.321431 0.025772 0.003780 0.023229 0.000000 彝族[13] 0.094028 0.189993 0.061012 0.054904 0.076758 0.022721 0.021975 0.327090 0.022979 0.008102 0.032636 0.008336 0.000000 纳西族[14] 0.094438 0.173688 0.058978 0.058992 0.078381 0.023124 0.018060 0.335448 0.024818 0.008774 0.023248 0.010194 0.009089 0.000000 日本人群[15] 0.099462 0.185518 0.066168 0.036783 0.080251 0.008683 0.010243 0.325122 0.026606 0.003187 0.023679 0.006106 0.008231 0.011813 0.000000 印度人群[16] 0.105637 0.186014 0.077436 0.059716 0.099815 0.024326 0.022901 0.334689 0.045724 0.014013 0.029487 0.014439 0.018083 0.019465 0.018190 0.000000 宁夏回族[17] 0.092766 0.179012 0.060018 0.053387 0.079086 0.017582 0.015580 0.320730 0.024201 0.004171 0.024065 0.003949 0.003815 0.006234 0.005949 0.011384 0.000000 伊犁维地区吾尔族[18] 0.011876 0.095765 0.100860 0.130743 0.108918 0.099432 0.099592 0.412676 0.091099 0.083543 0.096303 0.084121 0.084766 0.087187 0.084149 0.096855 0.081386 0.000000 长沙汉族[19] 0.050266 0.124632 0.097791 0.191280 0.091328 0.158492 0.153777 0.488916 0.125838 0.140880 0.136507 0.137303 0.142069 0.132549 0.147205 0.154349 0.138767 0.037496 0.000000 拉萨藏族[20] 0.019893 0.083069 0.118218 0.114822 0.127158 0.086276 0.087764 0.438257 0.095018 0.080479 0.087737 0.085672 0.089791 0.088243 0.082420 0.095327 0.084116 0.007435 0.053767 0.000000 怒江白族[21] 0.026784 0.091790 0.135895 0.142791 0.157151 0.111704 0.109450 0.448717 0.113373 0.097324 0.109013 0.103929 0.098482 0.099007 0.096941 0.117884 0.096831 0.014561 0.071961 0.014081 0.000000 大理白族[22] 0.015095 0.085721 0.112248 0.130195 0.122038 0.094348 0.096091 0.423142 0.096458 0.081886 0.090705 0.084250 0.082848 0.084821 0.083601 0.09195 0.07896 0.00521 0.04733 0.00502 0.01303 0.00000 -
[1] 张旭. 大理白族史探索[M]. 昆明: 云南人民出版社, 1990: 1-2. [2] Bra W,Brinkmann B,Budowle B,et al. DNA recommendations. Further report of the DNA Commission of the ISFH regarding the use of short tandem repeat systems. International Society for Forensic Haemogenetics[J]. Int J Legal Med,1997,110(4):175-176. doi: 10.1007/s004140050061 [3] Lincoln P J. DNA recommendations-further report of the DNA Commission of the ISFH regarding the use of short tandem repeat systems[J]. Forensic Sci Int,1997,87(3):181-184. [4] 赵方,伍新尧,蔡贵庆,等. Modified-Powerstates软件在法医生物统计中应用[J]. 中国法医学杂志,2003,18(5):297-298. doi: 10.3969/j.issn.1001-5728.2003.05.015 [5] Excoffier L,Lischer H E. Arlequin suite ver 3.5:A new series of programs to perform population genetics analyses under Linux and Windows[J]. Mol Ecol Resour,2010,10(3):564-567. doi: 10.1111/j.1755-0998.2010.02847.x [6] Kumar S,Stecher G,Tamura K. MEGA7:Molecular evolutionary genetics analysis Version 7.0 for bigger datasets[J]. Mol Biol Evol,2016,33(7):1870-1874. doi: 10.1093/molbev/msw054 [7] Fan H,Wang X,Ren Z,et al. Population data of 19 autosomal STR loci in the Li population from Hainan Province in southernmost China[J]. Int J Legal Med,2019,133(2):429-431. doi: 10.1007/s00414-018-1828-2 [8] 高波,邱平明. 新疆伊犁州维吾尔族20个STR基因座的遗传多态性[J]. 分子诊断与治疗杂志,2015,7(6):407-411. doi: 10.3969/j.issn.1674-6929.2015.06.009 [9] Guo F. Allele frequencies of 17 autosomal STR loci in the Va ethnic minority from Yunnan Province,Southwest China[J]. Int J Legal Med,2017,131(5):1251-1252. doi: 10.1007/s00414-017-1620-8 [10] Guo F. Genetic variation of 17 autosomal STR loci in the Dong ethnic minority from Guangxi Zhuang Autonomous Region,South China[J]. Int J Legal Med,2017,131(6):1537-1538. doi: 10.1007/s00414-017-1576-8 [11] Hashiyada M,Itakura Y,Nagashima T,et al. Polymorphism of 17 STRs by multiplex analysis in Japanese population[J]. Forensic Sci Int,2003,133(3):250-253. doi: 10.1016/S0379-0738(03)00075-6 [12] He G,Su Y,Zou X,et al. Allele frequencies of 15 autosomal STRs in Chinese Nakhi and Yi populations[J]. Int J Legal Med,2019,133(1):105-108. doi: 10.1007/s00414-018-1931-4 [13] He G,Wang M,Liu J,et al. Forensic features and phylogenetic analyses of Sichuan Han population via 23 autosomal STR loci included in the Huaxia Platinum System[J]. Int J Legal Med,2018,132(4):1079-1082. doi: 10.1007/s00414-017-1679-2 [14] Huang Y,Yao J,Li J,et al. Population genetic data for 17 autosomal STR markers in the Hani population from China[J]. Int J Legal Med,2015,129(5):995-996. doi: 10.1007/s00414-015-1176-4 [15] Kang L L,Zhao J,Liu K,et al. Allele frequencies of 15 STR loci of Tibetan lived in Xizang Lassa[J]. Forensic Sci Int,2007,168(2-3):236-240. doi: 10.1016/j.forsciint.2006.02.041Kang L L,Zhao J,Liu K,et al. Allele frequencies of 15 STR loci of Tibetan lived in Xizang Lassa[J]. Forensic Sci Int,2007,168(2-3):236-240. doi: 10.1016/j.forsciint.2006.02.041 [16] Li L,Zhang Y,Wang W,et al. Allele frequencies of 20 autosomal STR loci for 207 unrelated individuals of the Blang people in China[J]. Int J Legal Med,2020,134(3):987-988. doi: 10.1007/s00414-019-02192-0 [17] Li Y,Hong Y,Li X,et al. Allele frequency of 19 autosomal STR loci in the Bai population from the southwestern region of Mainland of China[J]. Electrophoresis,2015,36(19):2498-2503. doi: 10.1002/elps.201500129 [18] Li Z,Zhang J,Zhang H,et al. Genetic polymorphisms in 18 autosomal STR loci in the Tibetan population living in Xizang Chamdo,Southwest China[J]. Int J Legal Med,2018,132(3):733-734. doi: 10.1007/s00414-017-1740-1Li Z,Zhang J,Zhang H,et al. Genetic polymorphisms in 18 autosomal STR loci in the Tibetan population living in Xizang Chamdo,Southwest China[J]. Int J Legal Med,2018,132(3):733-734. doi: 10.1007/s00414-017-1740-1 [19] 刘亚举,李瑾,岳俊涛,等. 贵州仡佬族和苗族人群24个常染色体STR基因座的遗传多态性及遗传关系分析[J]. 解放军医学杂志,2019,44(8):682-689. doi: 10.11855/j.issn.0577-7402.2019.08.10 [20] Singh M,Nandineni M R. Population genetic analyses and evaluation of 22 autosomal STRs in Indian populations[J]. Int J Legal Med,2017,131(4):971-973. doi: 10.1007/s00414-016-1525-y [21] Sun J S,Tian Q H,Zhao L,et al. Genetic polymorphisms of 18 autosomal STR loci in Changsha Han population[J]. Journal of forensic medicine,2018,34(5):526-531. [22] 唐维,吴道来,陈立方,等. 云南白族15个常染色体STR基因座的遗传多态性[J]. 昆明医科大学学报,2016,37(5):128-131. doi: 10.3969/j.issn.1003-4706.2016.05.033 [23] Zhang J,Li Z,Mo X,et al. Allele frequencies of 18 autosomal STR loci in the Uyghur population living in Kashgar Prefecture,Northwest China[J]. Int J Legal Med,2019,133(2):427-428. doi: 10.1007/s00414-018-1821-9 [24] Zhang X,Hu L,Du L,et al. Genetic polymorphisms of 20 autosomal STR loci in the Vietnamese population from Yunnan Province,Southwest China[J]. Int J Legal Med,2017,131(3):661-662. doi: 10.1007/s00414-016-1496-z [25] Zhang X,Liu L,Xie R,et al. Population data and mutation rates of 20 autosomal STR loci in a Chinese Han population from Yunnan Province,Southwest China[J]. Int J Legal Med,2018,132(4):1083-1085. doi: 10.1007/s00414-017-1675-6 [26] 赵鹏,丁延云,张瑞智,等. 临夏回族人群15个STR多态性及与其他回族人群遗传关系初探[J]. 中国法医学杂志,2017,32(1):68-70. [27] 赖江华,张保华,朱波峰,等. 云南白族STR遗传多态性研究[J]. 西安交通大学学报(医学版),2002,23(3):242-245. doi: 10.3969/j.issn.1671-8259.2002.03.010 [28] 侯一平. 法医物证学[M]. 第4版. 北京: 人民卫生出版社, 2016: 190. [29] 中华人民共和国司法部. "亲权鉴定技术规范. "GB/T 37223-2018.2018-12-28. [30] 包睿舜,吴兰. 《中国孟高棉语族语言与南亚语系》评介[J]. 世界民族,2004,27(6):77-78. doi: 10.3969/j.issn.1006-8287.2004.06.011 [31] Cai X,Qin Z,Wen B,et al. Human migration through bottlenecks from Southeast Asia into East Asia during Last Glacial Maximum revealed by Y chromosomes[J]. PLoS One,2011,6(8):e24282. doi: 10.1371/journal.pone.0024282 [32] 戴庆厦,刘菊黄,傅爱兰. 关于我国藏缅语族系属分类问题[J]. 云南民族学院学报,1989,6(3):82-92. [33] Wang LX,Lu Y,Zhang C,et al. Reconstruction of Y-chromosome phylogeny reveals two neolithic expansions of Tibeto-Burman populations[J]. Mol Genet Genomics,2018,293(5):1293-1300. doi: 10.1007/s00438-018-1461-2 [34] 王嘉相. 勒墨人(白族支系)的迁徙与“指路歌”[A]. 政协怒江州委文史委. 怒江文史资料选辑[C]. 瑞丽: 德宏民族出版社, 1994: 63-65. -






 下载:
下载: