Analysis of Differential Gene Expression and Function in Peripheral Blood of Patients with Coronary Heart Disease and Acute Myocardial Infarction
-
摘要:
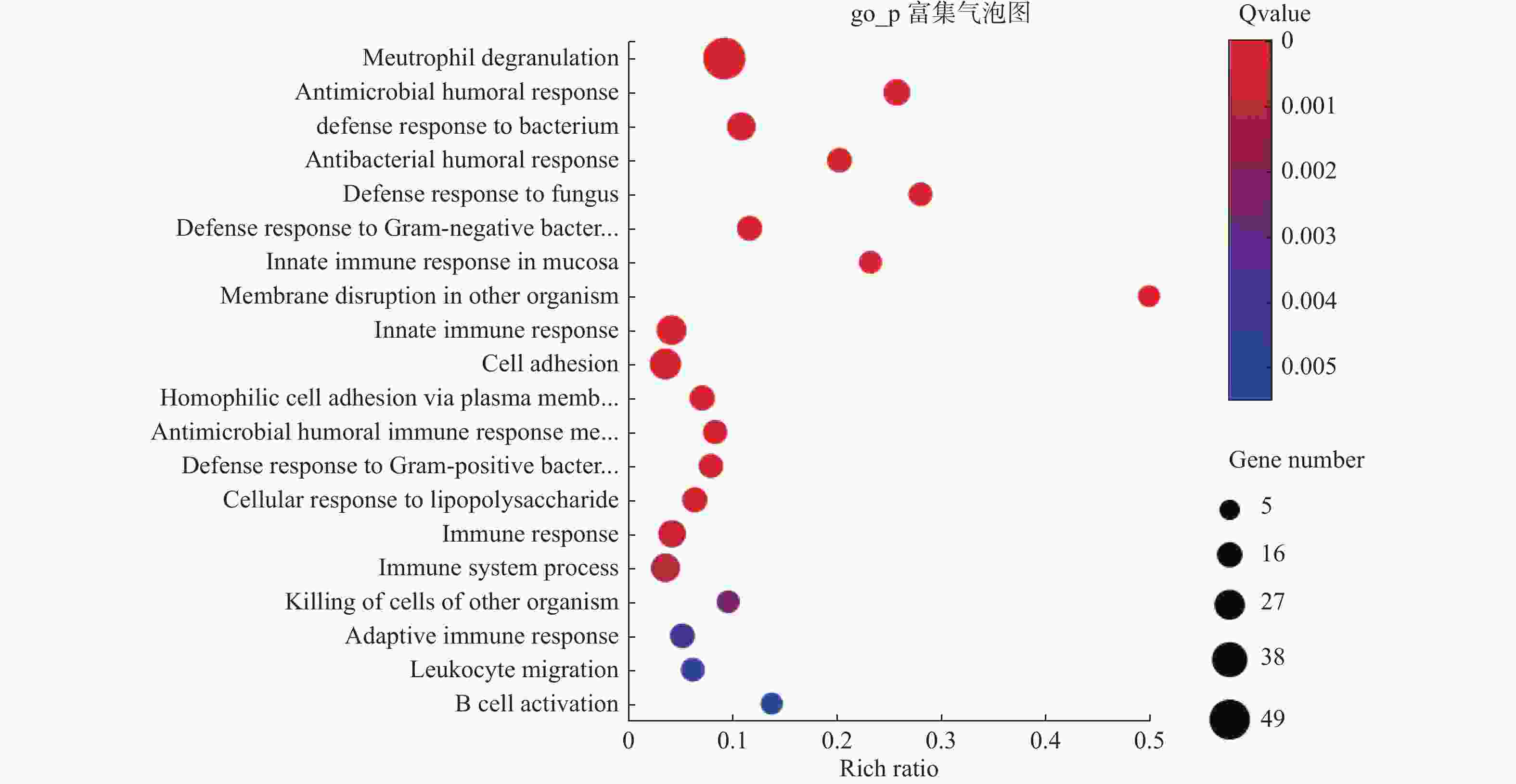
目的 探讨冠心病急性心肌梗死患者外周循环血单核细胞中差异基因表达及功能。 方法 收集2020年9月至2021 年9月在昆明市第一人民医院接受冠脉造影的患者,提取外周血单核细胞总RNA,使用DNBSEQ平台进行第2代高通量测序。检测并筛选差异表达基因,进行KEGG及GO富集分析。 结果 冠心病急性心肌梗死组与冠脉正常组对比,其中差异基因89个,上调基因67个,下调基因22个;其中差异lncRNA14个,差异mRNA72个,差异circRNA3个。完成KEGG pathway富集分析、KEGG disease富集分析、KEGG molecular富集分析。完成GO cell composition富集分析、GO cell function富集分析、GO biological process molecular富集分析。 结论 筛选出显著差异的10个mRNA、6个lncRNA、2个circRNA。富集分析中涉及感染、转录失调、PI3K-Akt信号通路、脂类合成、吞噬泡腔、细胞外基质、脂多糖结合等,与冠状动脉粥样硬化的发生与发展及急性血栓事件的发生可能相关。 Abstract:Objective To investigate the differential gene expression and function of peripheral blood monocytes in patients with coronary heart disease and acute myocardial infarction. Methods Patients receiving coronary angiography in Kunming First People’s Hospital from September 2020 to September 2021 were collected and total RNA of peripheral blood monocytes was extracted. DNBSEQ platform was used for second-generation high-throughput sequencing. We tested and screened differentially expressed genes, and made KEGG and GO enrichment analysis. Results There were 89 differentially expressed genes, 67 up-regulated genes and 22 down-regulated genes in acute myocardial infarction group compared with normal coronary artery group. Among them, there were 14 differential lncRNAs, 72 differential mRNAs, and 3 differential circRNAs. KEGG pathway enrichment analysis, KEGG disease enrichment analysis and KEGG molecular enrichment analysis were completed. GO cell composition enrichment analysis, GO cell function enrichment analysis and GO biological process molecular enrichment analysis were completed. Conclusions 10 mRNAs, 6 lncRNAs, and 2 circRNAs with significant differences were screened. The enrichment analysis involves infection, transcriptional disorder, PI3K Akt signaling pathway, lipid synthesis, phagocytic vesicle cavity, extracellular matrix, lipopolysaccharide binding, etc., which may be related to the occurrence and development of coronary atherosclerosis and the occurrence of acute thrombotic events. -
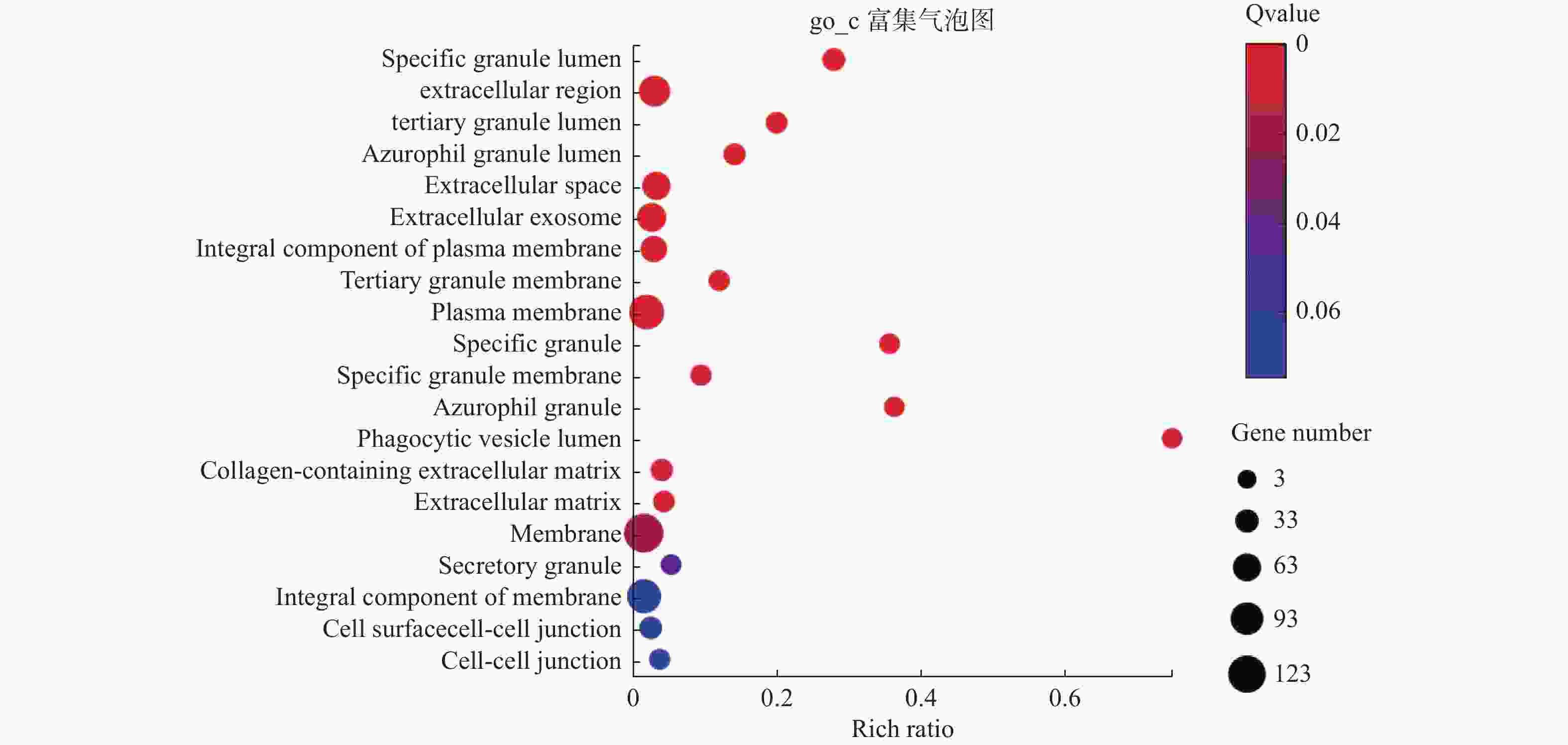
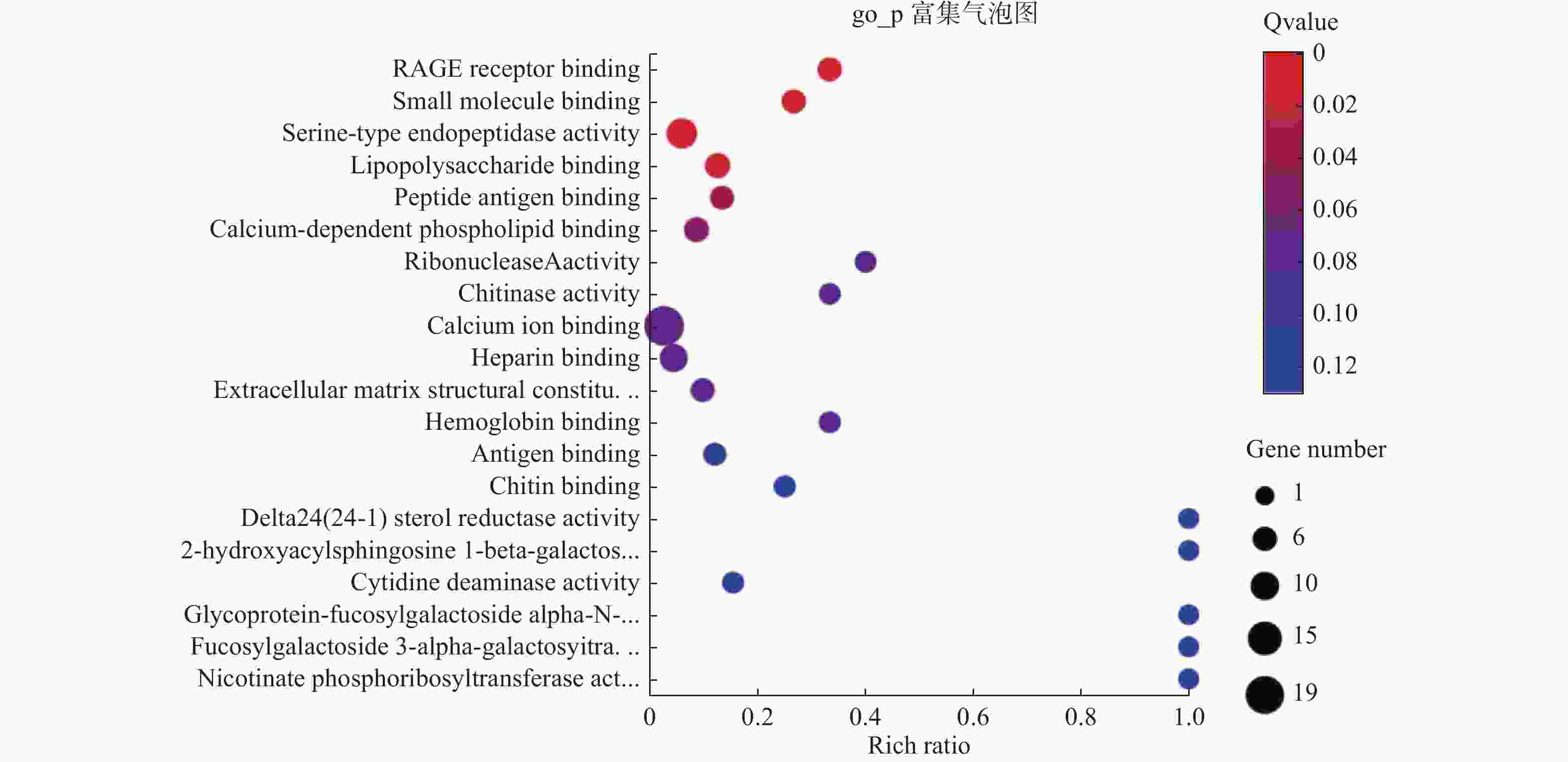
图 10 GO-CC富集分析
16个细胞组成显著富集分别为:specific granule lumen、extracellular region、tertiary granule lumen、azurophll granule lumen、ectracellular space、ectracellular exosome、integral component of plasma membrane、tertiary granule membrane、plasma membrane、specific granule、specific granule membrane、azurophil granule、phagocytic vesicle lumen、Collagen-containing extracellular matrix、extracellular matrix and membrane。
Figure 10. GO-cc enrichment analysis
表 1 4样本总RNA检测质控数据
Table 1. Total RNA test quality control data of 4 samples
样品名称 管数 浓度(ng/ μL) 体积(μL) 总量(μg) RIN 28S/ 18S 建库类型 结果 4A 1 195 40 7.8 9.5 2.0 DNBSEQ LncRNA-Seq 合格 5A 1 245 40 9.8 9.5 2.1 DNBSEQ LncRNA-Seq 合格 6A 1 430 40 17.2 9.3 2.1 DNBSEQ LncRNA-Seq 合格 7A 1 476 40 19.04 9.8 2.0 DNBSEQ LncRNA-Seq 合格 样本浓度及纯度均达质控标准。 表 2 过滤后质量(Reads过滤)
Table 2. Reads quality statistics after filtering (Reads filtering)
Sample name Total Raw
ReadsTotal Clean Reads Total Clean
BasesClean Reads Q20 Clean Reads Q30 Clean Reads
RatioCG5A 112.44 111.38 11.14 98.12 94.91 99.06 CG7A 109.94 109.18 10.92 97.95 94.59 99.30 EG4A 112.44 111.40 11.14 98.10 94.89 99.07 EG6A 109.94 109.20 10.92 97.90 94.45 99.32 样本reads达质控标准。 表 3 高通量测序4个样本患者基线资料
Table 3. Baseline data of 4 patients whose samples received high-throughput sequencing
项目 病变组 对照组 EG4A EG6A CG5A CG7A 年龄(岁) 68 57 83 56 性别 男 男 男 女 高血压 有/3级 有/3级 有/3级 有/3级 糖尿病 无 无 有 有 冠心病家族史 无 无 无 无 吸烟 无 有30年 无 无 饮酒 无 无 无 无 EF值(%) 69.7 65 62.9 67 糖化血红蛋白(%) 6.2 6.58 9.33 5.93 TC(mmol/L) 3.41 5.87 2.56 5.04 TG(mmol/L) 1.87 2.46 0.55 1.07 LDL-C(mmol/L) 2.17 3.99 1.04 3.37 Cr(μmol/L) 103.4 84.4 62.8 68.7 eGFR(mL/min/1.73 m2) 68 90 121 92 CRP(mg/L) 2.5 14.2 1.6 3.1 cTnT(μg/L) 0.506 0.613 0.025 0.004 Mb(μg/L) 493 886 51 25 CKMB(μg/L) 24.89 22.79 3.44 1.31 BNP(pg/mL) 19.17 385 203 183 PCT(ng/L) 0.03 0.26 0.04 0.08 IL-6(ng/mL) 4.4 4.5 3.8 4.8 WBC(109/L) 16.05 16.15 4.4 4.12 N(%) 87.4 83 51.3 61.9 N(109/L) 14.62 13.4 2.26 2.55 Hb(g/L) 152 144 189 142 PLT(109/L) 255 130 96 205 表 4 差异基因数量统计
Table 4. Number statistics of differential genes
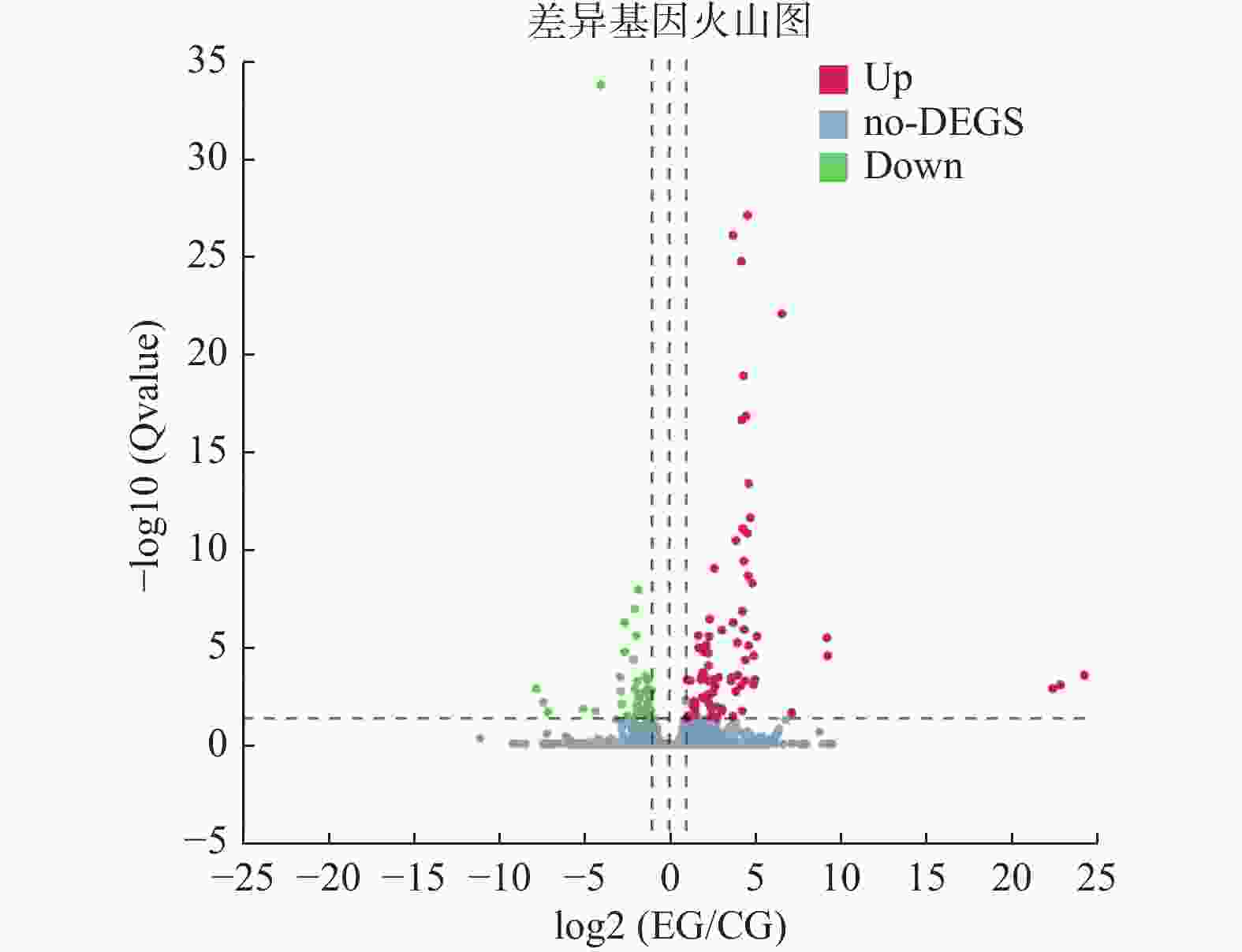
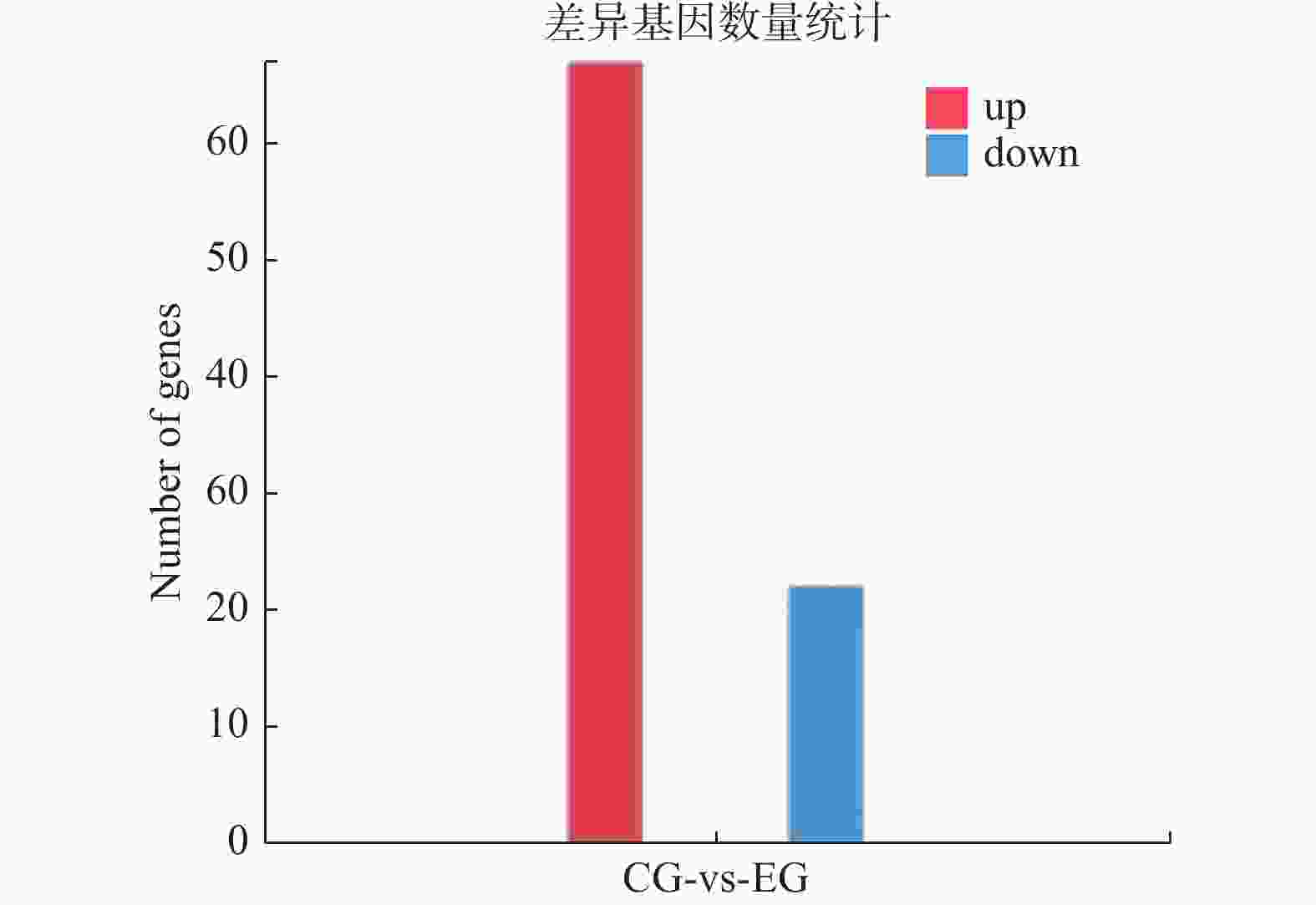
组别 下调 上调 总计 CG-vs-EG 22 67 89 EG(心梗组)与CG(对照组)对比差异基因共89个,其中上调基因67个,下调基因22个。 表 5 差异表达前10个mRNA
Table 5. Top 10 differentially expressed mRNA
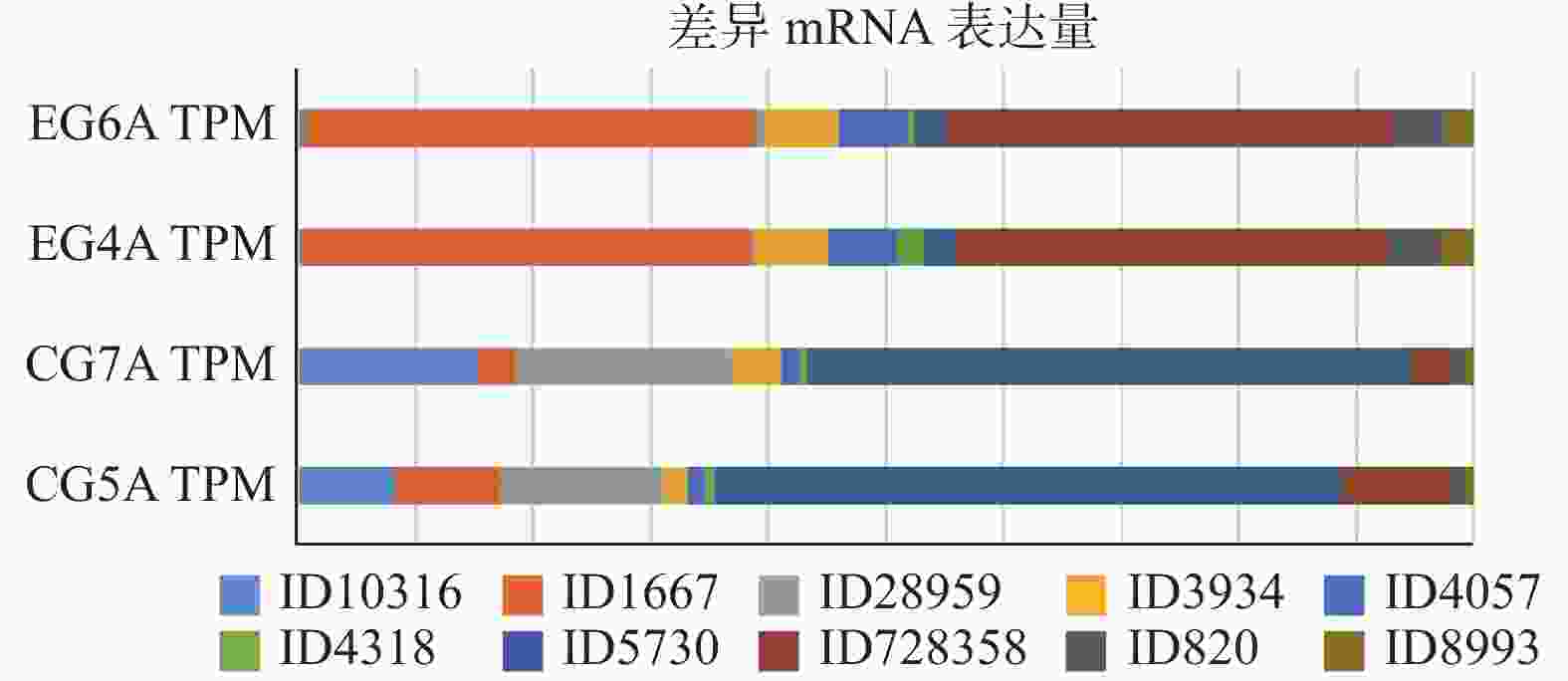
Gene ID Gene Symbol Chromosome CG5A TPM CG7A TPM EG4A TPM EG6A TPM P-value Q-value 上下调 10316 'NMUR1' chr2 67.92 77.05 16.85 15.04 < 0.001 < 0.001 Down 1667 'DEFA1' chr8 72.22 16.01 1378.42 1046.02 < 0.001 < 0.001 Up 28959 'TMEM176B' chr7 113.16 92.4 9.03 20.55 < 0.001 < 0.001 Down 3934 'LCN2' chr9 17.13 20.8 231.3 170.01 < 0.001 < 0.001 Up 4057 'LTF' chr3 11.18 7.47 208.24 158.71 < 0.001 < 0.001 Up 4318 'MMP9' chr20 7.79 3.76 84.47 15.73 < 0.001 < 0.001 Up 5730 'PTGDS' chr9 434.83 257.32 100.08 75.72 < 0.001 < 0.001 Down 728358 'DEFA1B' chr8 74.43 16.01 1334.87 1031.17 < 0.001 < 0.001 Up 820 'CAMP' chr3 10.12 7.74 152.14 123.63 < 0.001 < 0.001 Up 8993 'PGLYRP1' chr19 7.49 3.23 110.17 66.77 < 0.001 < 0.001 Up 差异表达前10个mRNA,其中上调mRNA7个,下调mRNA3个。 表 6 差异表达前6个lncRNA
Table 6. The top 6 differentially expressed lncRNA
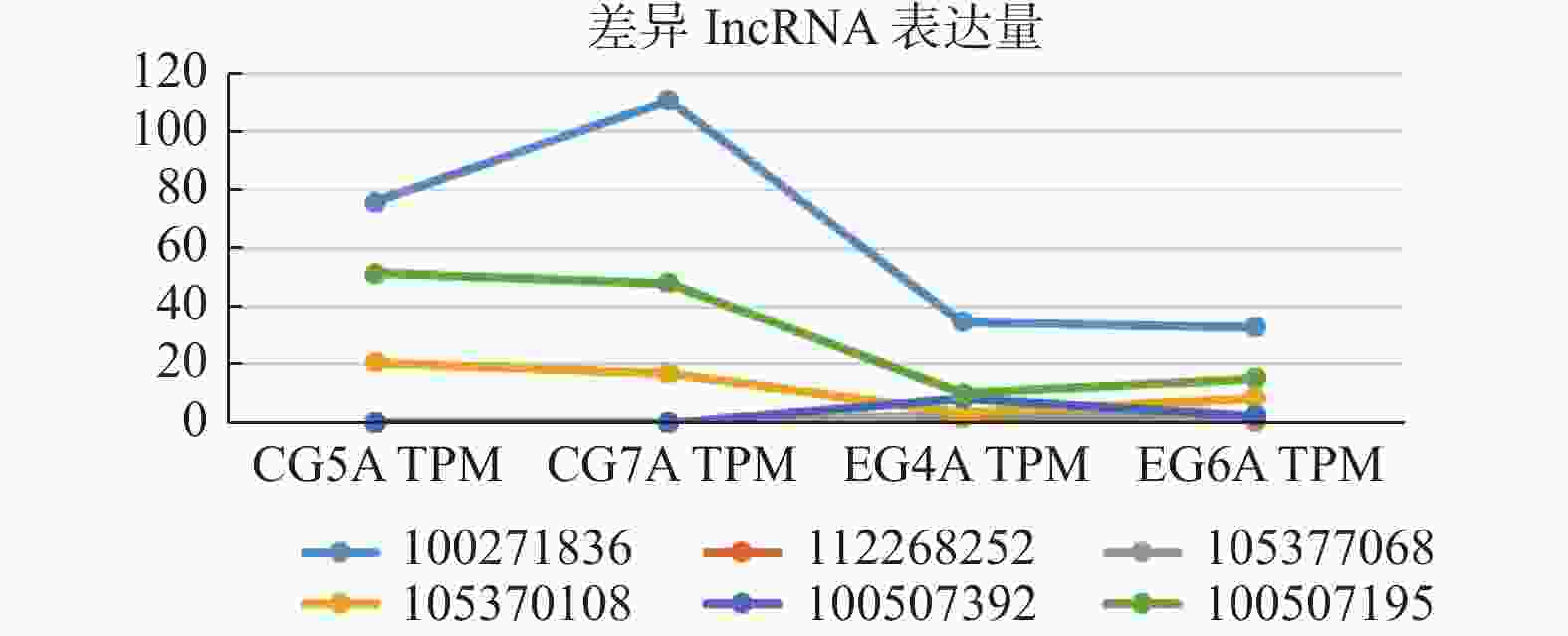
Gene ID Gene Symbol Chromosome CG5A TPM CG7A TPM EG4A TPM EG6A TPM P-value Q-value 上下调 100271836 'SMG1P3' chr16 75.69 110.78 34.65 32.74 < 0.001 0.001 Down 100507195 'LINC02384' chr12 51.41 48.19 10.17 15.29 < 0.001 < 0.001 Down 100507392 'SENCR' chr11 0 0 8.62 2.31 < 0.001 < 0.001 Up 105370108 'LOC105370108' chr13 20.63 17.07 3.38 8.49 < 0.001 0.036 Down 105377068 'LINC02009' chr3 0.07 0.15 2.79 2.29 < 0.001 < 0.001 Up 112268252 'LOC112268252' chr19 0.11 0.04 1.91 0.81 < 0.001 0.044 Up 差异表达前6个lncRNA,其中上调lncRNA3个,下调lncRNA3个。 表 7 差异表达前2个circRNA
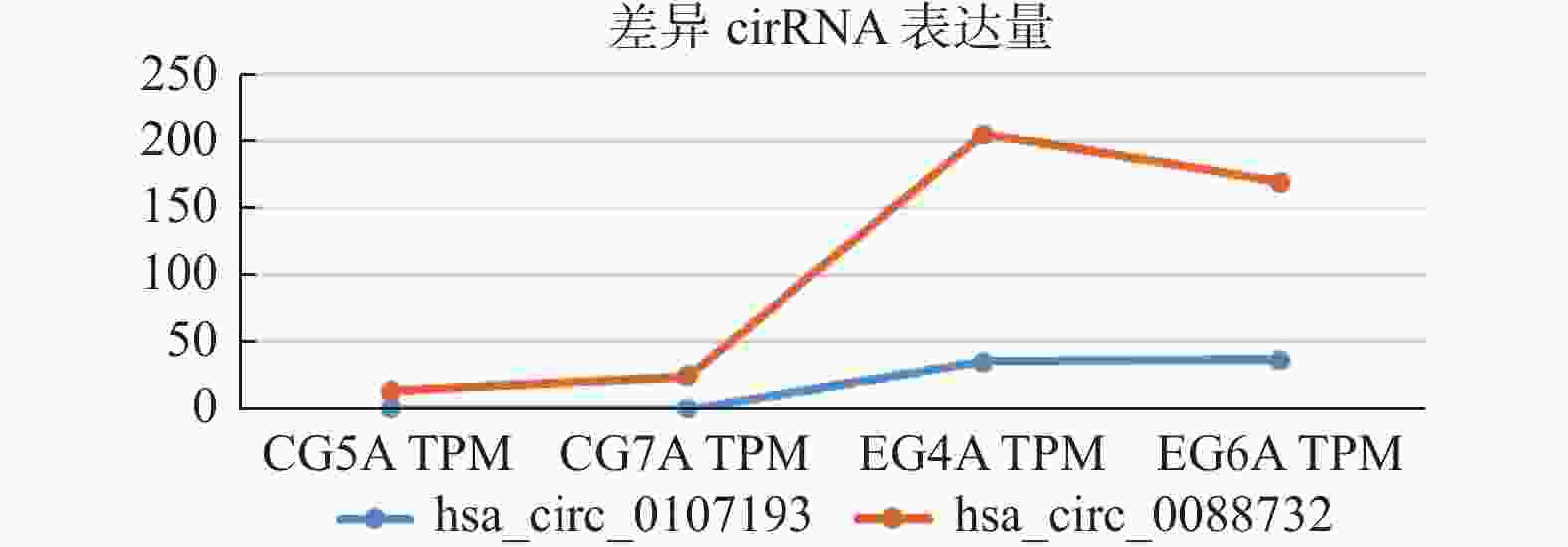
Table 7. The top 2 differentially expressed circRNA
Gene ID Chromosome CG5A TPM CG7A TPM EG4A TPM EG6A TPM P-value Q-value 上下调 hsa_circ_0088732 chr9 13.98004 24.87888 205.0836 169.3374 < 0.001 < 0.001 Up hsa_circ_0107193 chr17 0 0 35.81603901 36.51480767 < 0.001 < 0.001 Up 差异表达前2个circRNA,其中上调circRNA2个,下调circRNA 0个。 -
[1] 胡盛寿,高润霖,刘力生,等. 《中国心血管病报告2018》概要[J]. 中国循环杂志.,2019,34(3):209-220. [2] Thygesen K,Alpert JS,Jaffe AS,et al. Fourth Universal Definition of Myocardial Infarction (2018)[J]. Journal of the American College of Cardiology.,2018,72(18):2231-2264. doi: 10.1016/j.jacc.2018.08.1038 [3] Knorr M,Münzel T,Wenzel P. Interplay of NK cells and monocytes in vascular inflammation and myocardial infarction[J]. Front Physiol,2014,5(1):295. [4] Wirka RC,Pjanic M,Quertermous T. Advances in Transcriptomics:Investigating Cardiovascular Disease at Unprecedented Resolution[J]. Circ Res,2018,22(9):1200-1220. [5] GBD 2016 mortality collaborators. Global regional and national under-5 mortality adult mortality age specific mortality and lift expectancy 1970-2016:a systematic analysis for the Global Burden of Disease Study 2016[J]. Lancet,2017,390(10100):1084-1150. doi: 10.1016/S0140-6736(17)31833-0 [6] 世界卫生组织. 心血管疾病.https: //www.who.int/health-topics/cardiovascular-diseases, 2020-09-24. [7] Fabian Philipp Kreutzer,Jan Fiedler and Thomas Thum. Non-coding RNAs:key players in cardiac disease[J]. The journal of physiology.,2020,598(14):2995-3003. doi: 10.1113/JP278131 [8] Ballantyne M D,McDonald R A,Baker A H. lncRNA/microRNA interactions in the vasculature[J]. Clin Pharmacol Ther,2016,99(5):494-501. doi: 10.1002/cpt.355 [9] Jiangquan Liao,Jie Wang,Yongmei Liu,et al. Transcriptome sequencing of lncRNA,miRNA,mRNA and interaction network constructing in coronary heart disease[J]. BMC Medical Genomics,2019(12):124-136. [10] Salmena L,Poliseno L,Tay Y,et al. A ceRNA hypothesis:the Rosetta Stone of a hidden RNA language?[J]. Cell,2011,146(3):353-358. doi: 10.1016/j.cell.2011.07.014 [11] kugel J F,Goodrich J A. The regulation of mammalian mRNA transcription by lncRNAs:recent discoveries and current concepts[J]. Epigenomics,2013,5(2):95-102. [12] Han P,Chang C P. Long non-coding RNA and chromatin remodeling[J]. RNA Biol,2015,12(3):1094-1098. [13] Manduteanu I,Simionescu M. Inflammation in atherosclerosis:a cause or a result of vascular disorders?[J]. J Cell Mol Med,2012,16(7):1978-1990. [14] Hartge M M, Unger T, Kintscher U. The endothelium and vascular inflammation in diabetes[J]. Diab Vasc Dis Res 4(2): 84-88. [15] Wu G,Cai J,Han Y,et al. LincRNA –p21 regulates neointima formation,vascular smooth muscle cell proliferation,apoptosis,and atherosclerosis by enhancing p53 activity[J]. Circulation,2014,130(3):1452-1465. [16] Tang Y,Jin X,Xiang Y,et al. The lncRNA MALAT1 protects the endothelium against ox-LDL-induced dysfunction via upregulating the expression of the miR-22-3p target genes CXCR2 and AKT[J]. FEBS Lett,2015,589(6):3189-3196. [17] Feinberg M W. No small task:therapeutic targeting of Lp(a) for cardiovascular disease[J]. Lancet,2016,388(3):2211-2212. [18] Ping Li,Xinxin Yan,Guidong Xu,et al. A novel plasma lncRNA ENST00000416361 is upregulated in coronary artery disease and is related to inflammation and lipid metabolism[J]. Molecular Medicine Reports,2020,21(10):2375-2384. [19] Elling R,Chan J,and Fitzgerald K A. Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression[J]. Eur J Immunol,2016,46(1):504-512. [20] Lilly P,Lichtman A H,Hansson G K. Immune effector mechanisms implicated in atherosclerosis:from mice to humans[J]. Immunity,2013,38(2):1092-1104. [21] Chen R,Kong P,Zhang F,et al. EZH2-mediated alpha-actin methylation needs lncRNA TUG1 and promotes the cortex cytoskeleton formation in VSMCs[J]. Gene,2017,616(1):52-57. [22] Covarrubias S,Robinson E K,Shapleigh B,et al. CRISPR/Cas-based screening of long non-coding RNAs(lncRNAs)in macrophages with an NF-κB reporter[J]. Journal of Biological Chemistry,2017,292(12):20911-20920. -






 下载:
下载: