Genetic polymorphisms of 20 Autosomal STR loci in Miao Population from Wenshan and Honghe Regions, Yunnan Province
-
摘要:
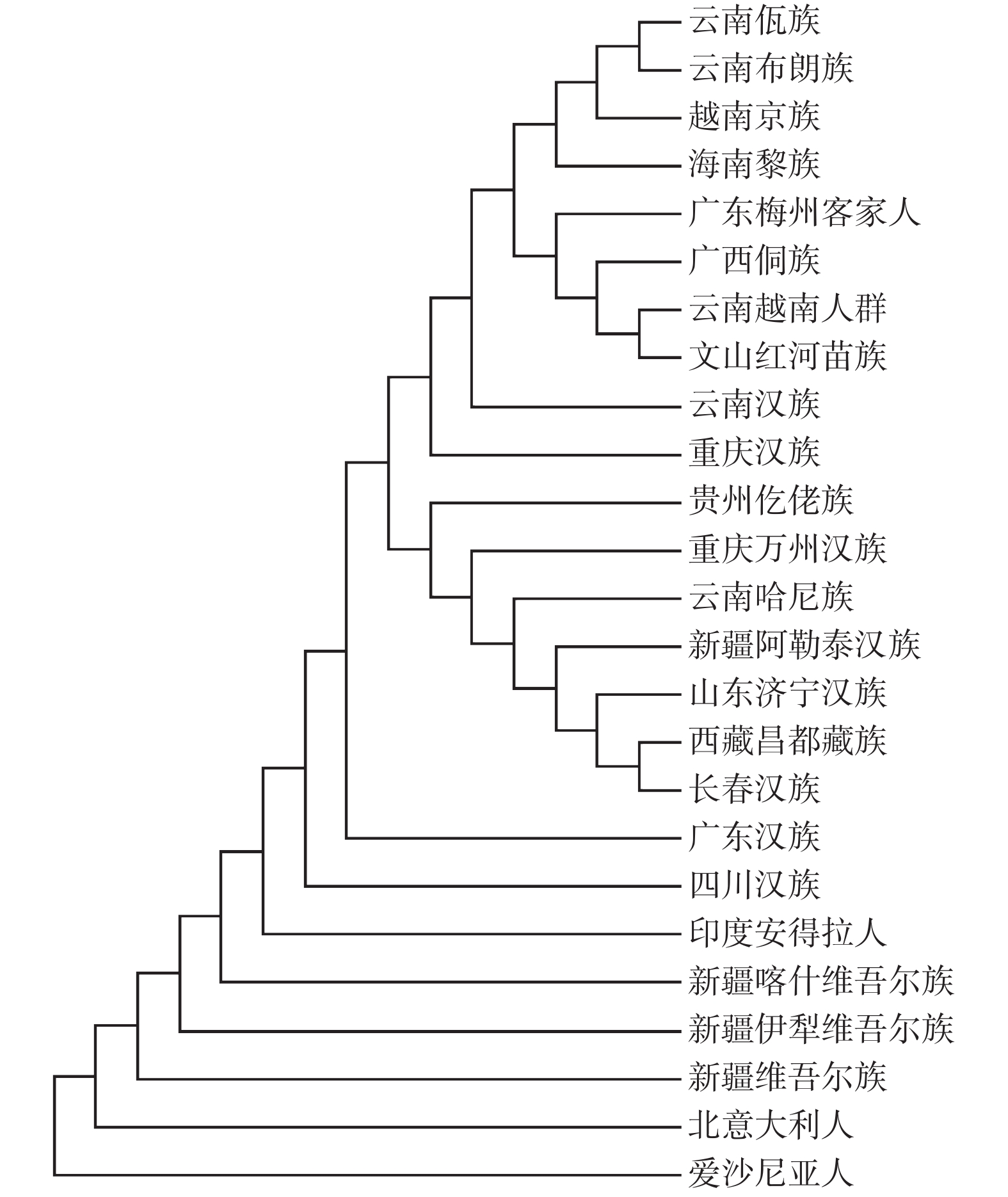
目的 研究文山红河地区苗族群体20个常染色体STR基因座的遗传多态性,评估其在该人群中的法医学应用价值,为法医DNA分析和人类遗传研究提供基础数据。 方法 采用Chelex-100法提取样本DNA,使用PowerPlex®21试剂盒对该地区2 209例苗族无关健康个体STR基因座进行复合扩增,用ABI 3130自动遗传分析仪对扩增产物进行毛细管电泳分析,用Genemapper ID v3.2软件进行基因分型。采用Modified-Powerstats软件进行Hardy-Weinberg平衡检验,并计算法医学参数。应用Arlequin和Phylip软件分别计算Fst值和Nei's遗传距离。使用SPSS软件进行多维尺度(MDS)分析。采用Mega软件构建相邻连接(N-J)系统发育树。 结果 20个常染色体STR基因座共检出265个等位基因和1080种基因型。等位基因频率分布为0.0002~0.5718,各基因座等位基因频率和基因型频率分布均符合Hardy-Weinberg平衡(P > 0.05/20 = 0.0025)。累积非父排除率(CPE)为 0.999 999 935 545 335,累积个人识别概率(CDP)为 0.999 999 999 999 999 999 999 932 674 151。Nei's遗传距离、N-J系统发育树和MDS分析显示,文山红河地区苗族与云南越南人群遗传关系最近。 结论 20个常染色体STR基因座在文山红河地区苗族群体中具有较高的遗传多态性,可用于法医学鉴定和群体遗传学研究。 Abstract:Objective To investigate the genetic polymorphisms of 20 autosomal STR loci in Miao population from Wenshan and Honghe regions, and to explore their forensic values. Methods DNA was extracted with the Chelex-100 method. A total of 20 STR loci were amplified from 2209 healthy unrelated individuals of the Miao population from Wenshan and Honghe regions, Yunnan Province using the PowerPlex®21 System kit. The PCR products were separated by capillary electrophoresis on an ABI 3130Genetic Analyzer, and alleles were genotyped by GeneMapper ID (Version 3.2) software. Forensic parameters were calculated by Modified-Powerstats software, and the Hardy-Weinberg equilibrium (HWE) test was performed. The Fst values and Nei's genetic distance were calculated by Arlequin and Phylip software, respectively. Multidimensional scaling (MDS) analysis was performed with SPSS v24.0 statistical software and the neighbor-joining (N-J) phylogenetic tree was constructed using the MEGA v6.0 software. Results A total of 265 alleles and 1080 genotypes were observed. The allele frequency of the 20 STRs ranged from 0.0002 to 0.5718. No deviation from Hardy-Weinberg equilibrium (P > 0.05/20 = 0.0025) was observed. The cumulative discrimination power (CDP) and cumulative probability of exclusion (CPE) with 20 STRs were 0.999 999 999 999 999 999 999 932 674 151 and 0.999 999 935 545 335, respectively. Population comparison showed that Wenshan and Honghe Miao nationality was genetically closer to Yunnan Vietnamese. Conclusion These 23 STR loci are highly genetic polymorphic and informative in the Miao population from Wenshan and Honghe regions, Yunnan Province, which can be used for forensic identification and population genetic research. -
表 1 文山红河地区苗族群体20个STR等位基因频率分布 (n = 2209) (1)
Table 1. Allele frequency distribution of 20 STR loci in Miao population from Wenshan and Honghe regions (n = 2209) (1)
D3S1358 CSF1PO TH01 TPOX vWA D13S317 FGA A F A F A F A F A F A F A F 13 0.0002 7 0.0007 5 0.001 7 0.0011 10 0.0002 7 0.0009 13 0.0005 14 0.0188 8 0.0002 6 0.1007 7.3 0.0002 11 0.0002 8 0.5718 13.2 0.0002 15 0.3279 9 0.0079 7 0.2831 8 0.3204 14 0.3268 9 0.0775 14 0.0002 16 0.2856 10 0.2156 8 0.0613 9 0.0840 15 0.0070 10 0.0965 14.2 0.0002 17 0.3202 11 0.1671 9 0.4755 10 0.0091 16 0.1231 11 0.1493 16 0.0005 18 0.0451 12 0.5634 9.8 0.0402 11 0.5471 17 0.1720 12 0.0634 18 0.0068 19 0.0023 13 0.0408 10 0.038 12 0.0374 18 0.3019 13 0.0333 19 0.0217 D1S1656 14 0.0043 12 0.0002 13 0.0007 19 0.0583 14 0.0066 20 0.0263 A F D8S1179 D5S818 Penta D 20 0.0104 15 0.0002 20.2 0.0002 10 0.0079 A F A F A F D7S820 18 0.0005 21 0.0625 11 0.0224 8 0.0034 7 0.0129 7 0.0041 A F D16S539 21.2 0.0011 12 0.0317 9 0.0005 8 0.0007 8 0.0568 7 0.0005 A F 22 0.0997 13 0.1135 10 0.3004 9 0.0222 9 0.3549 8 0.1928 8 0.0095 22.2 0.0020 14 0.1336 11 0.1453 10 0.2492 9.8 0.0002 9 0.0419 9 0.4380 23 0.0861 15 0.2455 12 0.0543 11 0.3115 10 0.1485 9.1 0.0009 10 0.0844 23.2 0.0054 16 0.2036 13 0.1335 12 0.2596 11 0.1225 10 0.1075 11 0.2150 24 0.2664 16.3 0.0072 14 0.0661 13 0.1363 11.2 0.0007 11 0.4855 12 0.2035 24.2 0.0430 17 0.0623 15 0.2417 14 0.0063 12 0.1661 12 0.1344 13 0.0290 25 0.2202 17.3 0.0476 16 0.0437 15 0.0005 13 0.0996 13 0.0179 14 0.0127 25.2 0.0229 18 0.0043 17 0.0068 29 0.0005 14 0.0455 14 0.0163 15 0.0075 25.4 0.0002 18.3 0.1098 18 0.0038 30 0.0005 15 0.0011 15 0.0023 16 0.0002 26 0.0945 19 0.0011 19 0.0005 19 0.0002 26.2 0.0027 19.3 0.0095 27 0.0344 28 0.0018 29 0.0002 A:等位基因,F:等位基因频率。 表 1 文山红河地区苗族群体20个STR等位基因频率分布 (n = 2209) (2)
Table 1. Allele frequency distribution of 20 STR loci in Miao population from Wenshan and Honghe regions (n = 2209) (2)
D2S1338 D18S51 D12S391 D21S11 D6S1043 Penta E D19S433 A F A F A F A F A F A F A F 13 0.0005 9 0.0002 10 0.0002 19 0.0002 8 0.0007 5 0.0337 9 0.0014 14 0.0005 10 0.0002 11 0.0002 21 0.0002 9 0.0088 8 0.0052 10 0.0007 16 0.0054 11 0.0011 14 0.0005 26 0.0002 10 0.0177 9 0.0018 10.2 0.0002 17 0.0380 12 0.0120 15 0.0054 27 0.0011 11 0.1225 10 0.0783 11 0.0005 18 0.0450 13 0.2130 15.2 0.0002 28 0.0222 12 0.0827 11 0.1139 11.2 0.0002 19 0.1668 14 0.2458 16 0.0002 28.2 0.0029 12.3 0.0002 11.4 0.0007 12 0.0222 20 0.0561 15 0.1446 17 0.0360 29 0.1966 13 0.1121 12 0.0645 12.2 0.0014 21 0.0217 16 0.2211 18 0.0829 30 0.2374 14 0.1298 13 0.0539 13 0.2584 22 0.0894 17 0.0269 19 0.3994 30.2 0.0079 14.3 0.0005 14 0.0876 13.2 0.1190 23 0.2836 18 0.0217 20 0.1097 30.3 0.0009 15 0.0179 15 0.1102 14 0.1446 24 0.2261 19 0.0607 21 0.1622 31 0.1271 17 0.0965 16 0.1220 14.2 0.0528 25 0.0383 20 0.0199 22 0.1382 31.2 0.1744 18 0.2249 17 0.0785 15 0.0311 26 0.0278 21 0.0093 23 0.0283 32 0.0129 18.2 0.0023 18 0.0781 15.2 0.3049 27 0.0007 22 0.0127 24 0.0195 32.2 0.0913 19 0.1175 19 0.0340 16 0.0091 23 0.0077 25 0.0138 33 0.0041 19.3 0.0018 20 0.0518 16.2 0.0199 24 0.0029 26 0.0032 33.2 0.1106 20 0.0405 21 0.0690 17 0.0311 34.2 0.0097 20.3 0.0106 22 0.0111 17.2 0.0009 21 0.0129 23 0.0020 18 0.0009 24 0.0002 24 0.0036 19 0.0002 25.2 0.0005 26 0.0002 A:等位基因,F:等位基因频率。 表 2 文山红河地区苗族人群20个STR基因座法医学参数 (n = 2209)
Table 2. Forensic parameters for 20 STR loci in Miao population from Wenshan and Honghe regions (n = 2209)
基因座 Pm DP PIC PE PIC Ho PHWE D3S1358 0.1417 0.8583 0.6483 0.4147 1.6188 0.6911 0.1202 D1S1656 0.0404 0.9596 0.8304 0.6773 3.1453 0.8410 0.3735 D6S1043 0.0289 0.9711 0.8597 0.7157 3.5844 0.8605 0.0875 D13S317 0.1659 0.8341 0.6011 0.3253 1.3425 0.6275 0.7826 Penta E 0.0132 0.9868 0.9108 0.8018 5.1612 0.9031 0.0167 D16S539 0.1239 0.8761 0.6702 0.4232 1.6485 0.6967 0.1026 D18S51 0.0575 0.9425 0.7943 0.6459 2.8540 0.8248 0.4664 D2S1338 0.0532 0.9468 0.8019 0.6081 2.5626 0.8049 0.0223 CSF1PO 0.2071 0.7929 0.5552 0.3156 1.3159 0.6200 0.1956 Penta D 0.0673 0.9327 0.7692 0.5855 2.4116 0.7927 0.8458 TH01 0.1629 0.8371 0.6202 0.3939 1.5491 0.6772 0.8096 vWA 0.1019 0.8981 0.7139 0.5282 2.0890 0.7607 0.4622 D21S11 0.0481 0.9519 0.8165 0.6860 3.2361 0.8455 0.2891 D7S820 0.1285 0.8715 0.6596 0.4358 1.6940 0.7048 0.3279 D5S818 0.1047 0.8953 0.7120 0.5240 2.0684 0.7583 0.6728 TPOX 0.2322 0.7678 0.5211 0.2576 1.1658 0.5711 0.0772 D8S1179 0.0664 0.9336 0.7767 0.6166 2.6235 0.8094 0.4679 D12S391 0.0759 0.9241 0.7488 0.5371 2.1344 0.7657 0.3763 D19S433 0.0679 0.9321 0.7731 0.5875 2.4242 0.7937 0.4834 FGA 0.0414 0.9586 0.8293 0.6985 3.3746 0.8518 0.4262 Pm:随机匹配概率;DP:个体识别概率;PIC:多态性信息含量;PE:非父排除概率;TPI:典型父权指数;Ho:观测值杂合度;PHWE:Hardy-Weinberg 平衡检验的P值。 表 3 文山红河苗族与24个参考群体的Fst值和P值(1)
Table 3. Pairwise Fst and P values between Wenshan and Honghe Miao and 24 reference populations(1)
基因座 新疆喀什维吾尔族 云南佤族 越南京族 印度安得拉人 广西侗族 海南黎族 西藏昌都藏族 广东汉族 CSF1PO Fst 0.04892 0.03811 0.03024 0.04620 0.02721 0.03571 0.02377 0.02719 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D13S317 Fst 0.12218 0.03381 0.04718 0.12030 0.03643 0.05175 0.24816 0.07356 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D16S539 Fst 0.04360 0.06322 0.07745 0.06543 0.02189 0.03740 0.04492 0.02776 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D18S51 Fst 0.01071 0.01880 0.01301 0.00989 0.01164 0.01761 0.01302 0.00857 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D19S433 Fst 0.04165 0.02227 0.02696 0.06136 0.02219 0.02905 0.03984 0.03079 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D21S11 Fst 0.01350 0.02236 0.01593 0.02074 0.01449 0.01474 0.01149 0.01519 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D2S1338 Fst 0.02820 0.02048 0.01664 0.02128 0.01498 0.02782 0.01408 0.01361 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D3S1358 Fst 0.01401 0.03336 0.00819 0.01508 0.00360 0.00342 0.28564 0.00935 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.00586 < 0.0001* < 0.0001* D5S818 Fst 0.01248 0.00881 0.00314 0.02870 0.00300 0.00572 0.00306 0.00379 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.03711 D7S820 Fst 0.03834 0.02294 0.01684 0.05025 0.01090 0.01764 0.03372 0.02488 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D8S1179 Fst 0.05130 0.04959 0.02108 0.04055 0.01420 0.03127 0.05009 0.03503 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* FGA Fst 0.02698 0.04601 0.03515 0.02453 0.03024 0.03777 0.02828 0.02921 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* TH01 Fst 0.04075 0.03605 0.02934 0.02730 0.01526 0.01895 0.02651 0.01834 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* TPOX Fst 0.26308 0.15765 0.11015 0.02832 0.07390 0.07432 0.12889 0.08064 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* vWA Fst 0.04013 0.21819 0.01131 0.04052 0.00897 0.00925 0.03436 0.01636 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* 经Bonferroni校正,*P < 0.05。 表 3 文山红河苗族与24个参考群体的Fst值和P值(2)
Table 3. Pairwise Fst and P values between Wenshan and Honghe Miao and 24 reference populations(2)
基因座 长春汉族 广东梅州客家人 云南哈尼族 爱沙尼亚人 新疆维吾尔族 新疆阿勒泰汉族 北意大利人 山东济宁汉族 CSF1PO Fst 0.02845 0.02576 0.03999 0.06563 0.06321 0.03096 0.07232 0.02451 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.13086 < 0.0001* D13S317 Fst 0.07338 0.07404 0.09289 0.18408 0.12976 0.06576 0.17795 0.07631 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D16S539 Fst 0.01968 0.02633 0.02399 0.07993 0.05339 0.02056 0.08816 0.01928 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.00488 < 0.0001* D18S51 Fst 0.00801 0.00740 0.02447 0.02643 0.01278 0.00889 0.02145 0.00944 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.17090 < 0.0001* D19S433 Fst 0.03042 0.02772 0.02379 0.08203 0.04816 0.03065 0.07360 0.03426 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D21S11 Fst 0.01518 0.01438 0.01283 0.02652 0.01642 0.01567 0.01655 0.01626 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D2S1338 Fst 0.01124 0.01391 0.01628 0.06878 0.02880 0.01358 0.06843 0.01282 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D3S1358 Fst 0.01494 0.00532 0.01147 0.02766 0.01634 0.00966 0.02401 0.01372 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.00391 < 0.0001* 0.70898 < 0.0001* D5S818 Fst 0.00548 0.00405 0.01370 0.02545 0.02061 0.00436 0.03307 0.00406 P < 0.0001* < 0.0001* < 0.0001* 0.22461 0.01855 < 0.0001* 0.50195 0.00098* D7S820 Fst 0.02726 0.01987 0.03182 0.07066 0.03906 0.02420 0.06307 0.03055 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.65137 < 0.0001* D8S1179 Fst 0.04940 0.03246 0.06440 0.07994 0.06182 0.04133 0.06670 0.04616 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.01660 < 0.0001* FGA Fst 0.02993 0.11697 0.03803 0.04585 0.02702 0.03103 0.04096 0.03085 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* TH01 Fst 0.02150 0.01648 0.03551 0.10167 0.05350 0.02031 0.11555 0.02209 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.00098* < 0.0001* TPOX Fst 0.07445 0.07704 0.13817 0.13248 0.10054 0.07168 0.09175 0.06013 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.08008 < 0.0001* vWA Fst 0.02037 0.01402 0.01448 0.04766 0.05542 0.01902 0.05278 0.02241 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.12207 < 0.0001* 经Bonferroni校正,*P < 0.05。 表 3 文山红河苗族与24个参考群体的Fst值和P值(3)
Table 3. Pairwise Fst and P values between Wenshan and Honghe Miao and 24 reference populations(3)
基因座 重庆汉族 新疆伊犁维吾尔族 云南汉族 贵州仡佬族 云南越南人群 重庆万州汉族 云南布朗族 四川汉族 CSF1PO Fst 0.03178 0.03343 0.03217 0.02091 0.00103 0.03921 0.03871 0.03047 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.13965 < 0.0001* < 0.0001* < 0.0001* D13S317 Fst 0.06456 0.13545 0.05557 0.06217 0.00683 0.06135 0.05160 0.06628 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D16S539 Fst 0.02318 0.05632 0.02659 0.03085 0.00468 0.02083 0.04630 0.01722 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.00098* < 0.0001* < 0.0001* < 0.0001* D18S51 Fst 0.00944 0.01731 0.00897 0.00461 0.00068 0.00821 0.00616 0.00901 P < 0.0001* < 0.0001* < 0.0001* 0.00293* 0.15430 < 0.0001* 0.00977 < 0.0001* D19S433 Fst 0.03200 0.04606 0.02251 0.02716 0.00115 0.02517 0.02561 0.02336 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.07812 < 0.0001* < 0.0001* < 0.0001* D21S11 Fst 0.01482 0.07764 0.01327 0.01314 0.00011 0.01627 0.01728 0.01565 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.33984 < 0.0001* < 0.0001* < 0.0001* D2S1338 Fst 0.01306 0.02377 0.01331 0.01143 0.00021 0.01356 0.02894 0.01220 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* < 0.0001* D3S1358 Fst 0.00722 0.02473 0.01089 0.00541 −0.00069 0.00579 0.01281 0.01077 P < 0.0001* < 0.0001* < 0.0001* 0.00879 0.73242 0.00391 0.00195* < 0.0001* D5S818 Fst 0.00339 0.01863 0.00141 0.00224 −0.00025 0.00293 0.00122 0.00356 P 0.00293* < 0.0001* 0.00781 0.05957 0.50781 0.02344 0.19824 0.02441 D7S820 Fst 0.01968 0.03992 0.01457 0.01572 −0.00058 0.01832 0.02008 0.02432 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.71582 < 0.0001* < 0.0001* < 0.0001* D8S1179 Fst 0.03540 0.06019 0.02853 0.03766 0.00229 0.03909 0.04517 0.03944 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.02832 < 0.0001* < 0.0001* < 0.0001* FGA Fst 0.02704 0.02258 0.02632 0.02894 0.00134 0.03556 0.03506 0.02807 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.04004 < 0.0001* < 0.0001* < 0.0001* TH01 Fst 0.02076 0.04799 0.02055 0.01987 0.00701 0.02384 0.04211 0.01968 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.00098* < 0.0001* < 0.0001* < 0.0001* TPOX Fst 0.07255 0.08357 0.08606 0.06064 0.00183 0.07700 0.15788 0.08363 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.07617 < 0.0001* < 0.0001* < 0.0001* vWA Fst 0.01477 0.04525 0.01162 0.01530 0.00092 0.01369 0.02023 0.02211 P < 0.0001* < 0.0001* < 0.0001* < 0.0001* 0.13477 < 0.0001* < 0.0001* < 0.0001* 经Bonferroni校正,P < 0.05。 表 4 文山红河苗族与24个参考群体的Nei's遗传距离
Table 4. Nei’s standard genetic distances between Wenshan and Honghe Miao and 24 reference populations
[1] [2] [3] [4] [5] [6] [7] [8] [9] [10] [11] [12] [13] [14] [15] [16] [17] [18] [19] [20] [21] [22] [23] [24] [25] 新疆喀什维吾尔族[1] 0.0000 云南佤族[2] 0.1208 0.0000 越南京族[3] 0.0449 0.0718 0.0000 印度安得拉人[4] 0.0343 0.1675 0.0855 0.0000 广西侗族[5] 0.0530 0.0841 0.0084 0.0803 0.0000 海南黎族[6] 0.0494 0.0860 0.0075 0.0816 0.0054 0.0000 西藏昌都藏族[7] 0.3302 0.4171 0.3396 0.3775 0.3251 0.3334 0.0000 广东汉族[8] 0.0343 0.0818 0.0122 0.0632 0.0091 0.0090 0.3012 0.0000 长春汉族[9] 0.0385 0.0954 0.0246 0.0649 0.0176 0.0173 0.2941 0.0035 0.0000 广东梅州客家人[10] 0.1071 0.1551 0.0715 0.1323 0.0691 0.0721 0.3958 0.0685 0.0746 0.0000 云南哈尼族[11] 0.0524 0.0866 0.0269 0.0893 0.0253 0.0245 0.3067 0.0130 0.0150 0.0767 0.0000 爱沙尼亚人[12] 0.7366 0.7277 0.7685 0.7730 0.8195 0.8001 1.5301 0.8232 0.8628 0.7329 0.8526 0.0000 新疆维吾尔族[13] 0.0061 0.1310 0.0551 0.0414 0.0685 0.0652 0.3466 0.0486 0.0550 0.1214 0.0709 0.7610 0.0000 新疆阿勒泰汉族[14] 0.0363 0.0903 0.0189 0.0661 0.0123 0.0125 0.2959 0.0020 0.0011 0.0720 0.0145 0.8411 0.0525 0.0000 北意大利人[15] 0.0381 0.2167 0.1308 0.0654 0.1447 0.1405 0.4020 0.1263 0.1377 0.2016 0.1427 0.7182 0.0371 0.1314 0.0000 山东济宁汉族[16] 0.0399 0.0963 0.0253 0.0656 0.0187 0.0177 0.2967 0.0039 0.0008 0.0759 0.0159 0.8590 0.0565 0.0014 0.1392 0.0000 重庆汉族[17] 0.0408 0.0799 0.0123 0.0680 0.0075 0.0079 0.3084 0.0020 0.0056 0.0705 0.0157 0.8155 0.0558 0.0030 0.1382 0.0057 0.0000 新疆伊犁维吾尔族[18] 0.0091 0.1246 0.0534 0.0423 0.0662 0.0613 0.3416 0.0451 0.0528 0.1207 0.0622 0.7705 0.0091 0.0505 0.0433 0.0535 0.0520 0.0000 云南汉族[19] 0.0385 0.0798 0.0103 0.0707 0.0071 0.0087 0.3075 0.0021 0.0051 0.0668 0.0145 0.8179 0.0540 0.0030 0.1352 0.0060 0.0023 0.0513 0.0000 贵州仡佬族[20] 0.0416 0.0927 0.0171 0.0697 0.0087 0.0097 0.3048 0.0037 0.0050 0.0692 0.0204 0.8291 0.0589 0.0028 0.1393 0.0053 0.0032 0.0560 0.0034 0.0000 云南越南人群[21] 0.0964 0.1554 0.0615 0.0987 0.0445 0.0517 0.3965 0.0583 0.0664 0.1081 0.0899 0.8656 0.1149 0.0597 0.1890 0.0642 0.0527 0.1127 0.0540 0.0511 0.0000 重庆万州汉族[22] 0.0465 0.0902 0.0160 0.0792 0.0093 0.0101 0.3036 0.0044 0.0051 0.0720 0.0151 0.8302 0.0646 0.0028 0.1457 0.0055 0.0045 0.0627 0.0040 0.0033 0.0567 0.0000 云南布朗族[23] 0.0515 0.0650 0.0112 0.1003 0.0178 0.0222 0.3235 0.0189 0.0322 0.0816 0.0280 0.7655 0.0605 0.0263 0.1340 0.0331 0.0186 0.0580 0.0176 0.0255 0.0825 0.0251 0.0000 四川汉族[24] 0.0347 0.0852 0.0171 0.0640 0.0145 0.0146 0.3015 0.0030 0.0041 0.0747 0.0151 0.8182 0.0493 0.0031 0.1297 0.0046 0.0039 0.0451 0.0037 0.0061 0.0635 0.0065 0.0234 0.0000 文山红河苗族[25] 0.1188 0.1768 0.0788 0.1151 0.0546 0.0697 0.4233 0.0750 0.0806 0.1197 0.1114 0.9045 0.1402 0.0741 0.2244 0.0795 0.0687 0.1377 0.0661 0.0633 0.0265 0.0715 0.1036 0.0765 0.0000 -
[1] 侯一平. 法医物证学[M]. 第4版. 北京: 人民卫生出版社, 2016: 32-33. [2] 谢波,杨平,余政梁,等. 多系统联合分析在复杂亲缘关系鉴定中的应用评估[J]. 中国法医学杂志,2021,36(2):173-176,180. doi: 10.13618/j.issn.1001-5728.2021.02.012 [3] Steffen C R,Huszar T I,Borsuk L A,et al. A multi-dimensional evaluation of the 'NIST 1032' sample set across four forensic Y-STR multiplexes[J]. Forensic Sci Int Genet,2022,57:102655. [4] 云南省统计局. 2021云南统计年鉴[M]. 北京: 中国统计出版社, 2021. [5] Excoffier L,Lischer H E. Arlequin suite ver 3.5:a new series of programs to perform population genetics analyses under Linux and Windows[J]. Mol Ecol Resour,2010,10(3):564-567. doi: 10.1111/j.1755-0998.2010.02847.x [6] Bindu G H,Trivedi R,Kashyap V K. Allele frequency distribution based on 17 STR markers in three major Dravidian linguistic populations of Andhra Pradesh,India[J]. Forensic Sci Int,2007,170(1):76-85. doi: 10.1016/j.forsciint.2006.04.012 [7] Chen P,Wang B,Gao B,et al. Forensic features and genetic structure of 23 autosomal STRs in Artux Turkic-speaking population residing in southwestern Xinjiang Uyghur Autonomous Region[J]. Int J Legal Med,2019,133(5):1393-1395. doi: 10.1007/s00414-019-02072-7 [8] Dang Z,Liu Q,Zhang G,et al. Population genetic data from 23 autosomal STR loci of Huaxia Platinum system in the Jining Han population[J]. Mol Genet Genomic Med,2020,8(4):e1142. [9] Fan H,Wang X,Ren Z,et al. Population data of 19 autosomal STR loci in the Li population from Hainan Province in southernmost China[J]. Int J Legal Med,2019,133(2):429-431. doi: 10.1007/s00414-018-1828-2 [10] Feng Z,Xia M,Bao H,et al. Genetic polymorphisms of 18 short tandem repeat loci in 3550 individuals from the Han population of Changchun,Northeast China[J]. Int J Legal Med,2016,130(6):1481-1483. doi: 10.1007/s00414-016-1339-y [11] 高波,邱平明. 新疆伊犁州维吾尔族20个STR基因座的遗传多态性[J]. 分子诊断与治疗杂志,2015,7(6):407-411. doi: 10.3969/j.issn.1674-6929.2015.06.009 [12] Guo F. Allele frequencies of 17 autosomal STR loci in the Va ethnic minority from Yunnan Province,Southwest China[J]. Int J Legal Med,2017,131(5):1251-1252. doi: 10.1007/s00414-017-1620-8 [13] Guo F. Genetic variation of 17 autosomal STR loci in the Dong ethnic minority from Guangxi Zhuang Autonomous Region,South China[J]. Int J Legal Med,2017,131(6):1537-1538. doi: 10.1007/s00414-017-1576-8 [14] Han X,Shen A,Yao T,et al. Genetic diversity of 17 autosomal STR loci in Meizhou Hakka population[J]. Int J Legal Med,2021,135(2):443-444. doi: 10.1007/s00414-020-02253-9 [15] He G,Wang M,Liu J,et al. Forensic features and phylogenetic analyses of Sichuan Han population via 23 autosomal STR loci included in the Huaxia Platinum System[J]. Int J Legal Med,2018,132(4):1079-1082. doi: 10.1007/s00414-017-1679-2 [16] Huang Y,Yao J,Li J,et al. Population genetic data for 17 autosomal STR markers in the Hani population from China[J]. Int J Legal Med,2015,129(5):995-996. doi: 10.1007/s00414-015-1176-4 [17] Li L,Zhang Y,Wang W,et al. Allele frequencies of 20 autosomal STR loci for 207 unrelated individuals of the Blang people in China[J]. Int J Legal Med,2020,134(3):987-988. doi: 10.1007/s00414-019-02192-0 [18] Li X,Li L,Wang Q,et al. Population genetic analysis of the Globalfiler STR loci in 3032 individuals from the Altay Han population of Xinjiang in northwest China[J]. Int J Legal Med,2018,132(1):141-143. doi: 10.1007/s00414-017-1641-3 [19] Li Z,Zhang J,Zhang H,et al. Genetic polymorphisms in 18 autosomal STR loci in the Tibetan population living in Xizang Chamdo,Southwest China[J]. Int J Legal Med,2018,132(3):733-734. doi: 10.1007/s00414-017-1740-1Li Z,Zhang J,Zhang H,et al. Genetic polymorphisms in 18 autosomal STR loci in the Tibetan population living in Xizang Chamdo,Southwest China[J]. Int J Legal Med,2018,132(3):733-734. doi: 10.1007/s00414-017-1740-1 [20] 刘亚举,李瑾,岳俊涛,等. 贵州仡佬族和苗族人群24个常染色体STR基因座的遗传多态性及遗传关系分析[J]. 解放军医学杂志,2019,44(8):682-689. [21] Sadam M,Tasa G,Tiidla A,et al. Population data for 22 autosomal STR loci from Estonia[J]. Int J Legal Med,2015,129(6):1219-1220. doi: 10.1007/s00414-014-1089-7 [22] Tran H L,Nguyen H T,Pham T T,et al. Allele frequencies for 22 autosomal STRs in the Kinh population in Vietnam[J]. Int J Legal Med,2019,133(6):1761-1762. doi: 10.1007/s00414-018-01996-w [23] Turrina S,Ferrian M,Caratti S,et al. Evaluation of genetic parameters of 22 autosomal STR loci (PowerPlex® Fusion System) in a population sample from Northern Italy[J]. Int J Legal Med,2014,128(2):281-283. doi: 10.1007/s00414-013-0934-4 [24] Yang L,Zhang X,Zhao L,et al. Population data of 23 autosomal STR loci in the Chinese Han population from Guangdong Province in southern China[J]. Int J Legal Med,2018,132(1):133-135. doi: 10.1007/s00414-017-1588-4 [25] Zhang J,Li Z,Mo X,et al. Allele frequencies of 18 autosomal STR loci in the Uyghur population living in Kashgar Prefecture,Northwest China[J]. Int J Legal Med,2019,133(2):427-428. doi: 10.1007/s00414-018-1821-9 [26] Zhang X,Hu L,Du L,et al. Genetic polymorphisms of 20 autosomal STR loci in the Vietnamese population from Yunnan Province,Southwest China[J]. Int J Legal Med,2017,131(3):661-662. doi: 10.1007/s00414-016-1496-z [27] Zhang X,Liu L,Xie R,et al. Population data and mutation rates of 20 autosomal STR loci in a Chinese Han population from Yunnan Province,Southwest China[J]. Int J Legal Med,2018,132(4):1083-1085. doi: 10.1007/s00414-017-1675-6 [28] Zou X,Li Y,Li P,et al. Genetic polymorphisms for 19 autosomal STR loci of Chongqing Han ethnicity and phylogenetic structure exploration among 28 Chinese populations[J]. Int J Legal Med,2017,131(6):1539-1542. doi: 10.1007/s00414-017-1577-7 [29] Zou X,Li Y,Wei Z,et al. Population data and forensic efficiency of 21 autosomal STR loci included in AGCU EX22 amplification system in the Wanzhou Han population[J]. Int J Legal Med,2018,132(1):153-155. doi: 10.1007/s00414-017-1680-9 [30] Kumar S,Stecher G,Tamura K. MEGA7:Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets[J]. Mol Biol Evol,2016,33(7):1870-1874. doi: 10.1093/molbev/msw054 [31] 李和. 古代云南文山地区与越南关系论述[J]. 文山学院学报,2013,26(2):64-69. doi: 10.3969/j.issn.1674-9200.2013.02.013 [32] 方玲,胡启平,潘尚领,等. 广西壮、苗族和越南京族人线粒体DNA 9 bp缺失频率的分析[J]. 国际遗传学杂志,2007,30(5):329-331. doi: 10.3760/cma.j.issn.1673-4386.2007.05.003 [33] 孙灏琳,李威,沈淼淼,等. 通过常染色体STR基因座进行两两族源推断初探[J]. 中国法医学杂志,2022,37(4):345-351. doi: 10.13618/j.issn.1001-5728.2022.04.006 -






 下载:
下载: