Genetic Polymorphisms of 17 Autosomal STR Loci in the Jino Population of Yunnan
-
摘要:
目的 对中国云南省基诺族人群无关个体进行 17个 STR 基因座遗传多态性调查,评估数据在人群中的法医学应用价值。 方法 检验331例基诺族无关个体,用AGCU 17+1试剂盒进行STR基因型扩增检测,使用ABI3130毛细管电泳仪分离扩增产物片段并进行等位基因分型。 结果 共检出186个等位基因,等位基因频率分布在 0.0015 ~0.5619 ,TPOX、TH01基因座多态性和识别能力最低,D3S1358、D13S317、D16S539、CSF1PO、D5S818均有参数显示多态性不佳,其他基因座显示有较好的多态性。基因型分布均符合哈温平衡,CDP和CPE均显示有较好的识别度。结论 除TPOX和TH01基因座,其余的15个常染色体STR 基因座在基诺族群体中具有较好的多态性,可以满足法医个体识别和亲权鉴定应用。 Abstract:Objective To study the genetic polymorphism of 17 autosomal STR loci in the Jino people of Yunnan, and to evaluate its value in forensic medicine. Methods A total of 331 unrelated individuals of the Jino people in Yunnan Province were examined. AGCU 17+1 kit was used for STR genotyping, and ABI3130 capillary electrophoresis apparatus was used to isolate the amplified product fragments and to determineallele type . Results A total of 186 alleles were detected, and the allele frequencies ranged from 0.0015 to0.561 9 . TPOX and TH01 loci had the lowest polymorphism and recognition ability, and D3S1358, D13S317, D16S539, CSF1PO and D5S818 had poor polymorphism parameters. Other loci showed better polymorphism. The distribution of genotypes was in accordance with Hardy-Weinberg equilibrium, and CDP and CPE showed good recognition.Conclusion Except for TPOX and TH01, the remaining 15 autosomal STR loci have good polymorphism in Jino population, which can meet the application of forensic individual identification and paternity identification. Except the TPOX and TH01 loci, the remaining 15 autosomal STR loci have good polymorphisms in the Jino population. -
Key words:
- Forensic physical evidence /
- Jino Population /
- Short tandem repeats
-
表 1 基诺族群体 17 个 STR 等位基因频率分布(n = 331)
Table 1. Frequency of 17 STR loci in the Jino people (n = 331)
D6S1043 D21S11 D18S51 FGA PENTA_E A F A F A F A F A F 10 0.015106 27 0.001511 10 0.001511 18 0.095166 5 0.030211 11 0.194864 28 0.015106 11 0.003021 19 0.021148 8 0.001511 12 0.252266 28.2 0.003021 12 0.016616 20 0.031722 10 0.036254 13 0.125378 29 0.228097 13 0.212991 21 0.197885 11 0.271903 14 0.101208 30 0.305136 14 0.240181 21.2 0.007553 12 0.108761 15 0.006042 30.2 0.031722 15 0.146526 22 0.172205 13 0.057402 16 0.003021 30.3 0.001511 16 0.116314 22.2 0.019637 14 0.148036 17 0.022659 31 0.086103 17 0.055891 23 0.140483 15 0.048338 17.2 0.003021 31.2 0.093656 18 0.034743 23.2 0.021148 16 0.058912 18 0.10423 32 0.016616 19 0.061934 24 0.163142 17 0.045317 19 0.099698 32.1 0.003021 20 0.043807 24.2 0.003021 18 0.089124 20 0.054381 32.2 0.155589 21 0.012085 25 0.089124 19 0.05287 20.3 0.003021 33 0.009063 22 0.009063 25.2 0.003021 20 0.013595 21 0.007553 33.2 0.049849 23 0.019637 26 0.024169 21 0.02719 21.3 0.007553 24 0.016616 27 0.007553 22 0.009063 25 0.006042 28 0.001511 23 0.001511 D19S433 D2S1338 26 0.003021 28.2 0.001511 A F A F CSF1PO 9 0.006042 16 0.033233 TPOX VWA A F 12 0.048338 16.1 0.001511 A F A F 7 0.006042 12.2 0.004532 17 0.092145 7 0.003021 12 0.004532 8 0.001511 13 0.279456 18 0.069486 8 0.561934 12.2 0.001511 9 0.013595 13.2 0.055891 19 0.259819 9 0.152568 13 0.003021 10 0.267372 14 0.291541 20 0.148036 10 0.013595 14 0.350453 11 0.280967 14.2 0.13142 21 0.028701 11 0.26284 15 0.022659 11.1 0.003021 15 0.039275 22 0.013595 12 0.006042 16 0.1571 12 0.367069 15.2 0.101208 23 0.101208 17 0.238671 13 0.02719 16 0.001511 24 0.135952 D3S1358 18 0.141994 14 0.033233 16.2 0.036254 25 0.099698 A F 19 0.064955 17.2 0.003021 26 0.010574 14 0.070997 20 0.012085 D13S317 18.2 0.001511 28 0.006042 15 0.358006 21 0.001511 A F 16 0.279456 23 0.001511 8 0.433535 D7S820 D8S1179 17 0.259819 9 0.07855 A F A F 18 0.030211 10 0.13142 8 0.125378 10 0.111782 19 0.001511 TH01 11 0.250755 9 0.151057 11 0.110272 A F 12 0.084592 9.1 0.001511 12 0.199396 D5S818 5 0.001511 13 0.009063 10 0.146526 13 0.223565 A F 6 0.070997 14 0.009063 11 0.320242 14 0.172205 7 0.003021 7 0.261329 15 0.003021 11.3 0.001511 15 0.135952 9 0.081571 8 0.075529 12 0.217523 16 0.024169 10 0.188822 8.3 0.003021 D16S539 13 0.02719 17 0.022659 11 0.194864 9 0.483384 A F 14 0.009063 12 0.392749 9.3 0.08006 8 0.001511 13 0.132931 10 0.024169 9 0.323263 13.2 0.001511 10 0.083082 14 0.004532 11 0.276435 12 0.222054 13 0.083082 14 0.010574 表 2 基诺族人群17个 STR 基因座法医学参数(n = 331)
Table 2. Forensic parameters of 17 STR loci in the Jino people (n = 331)
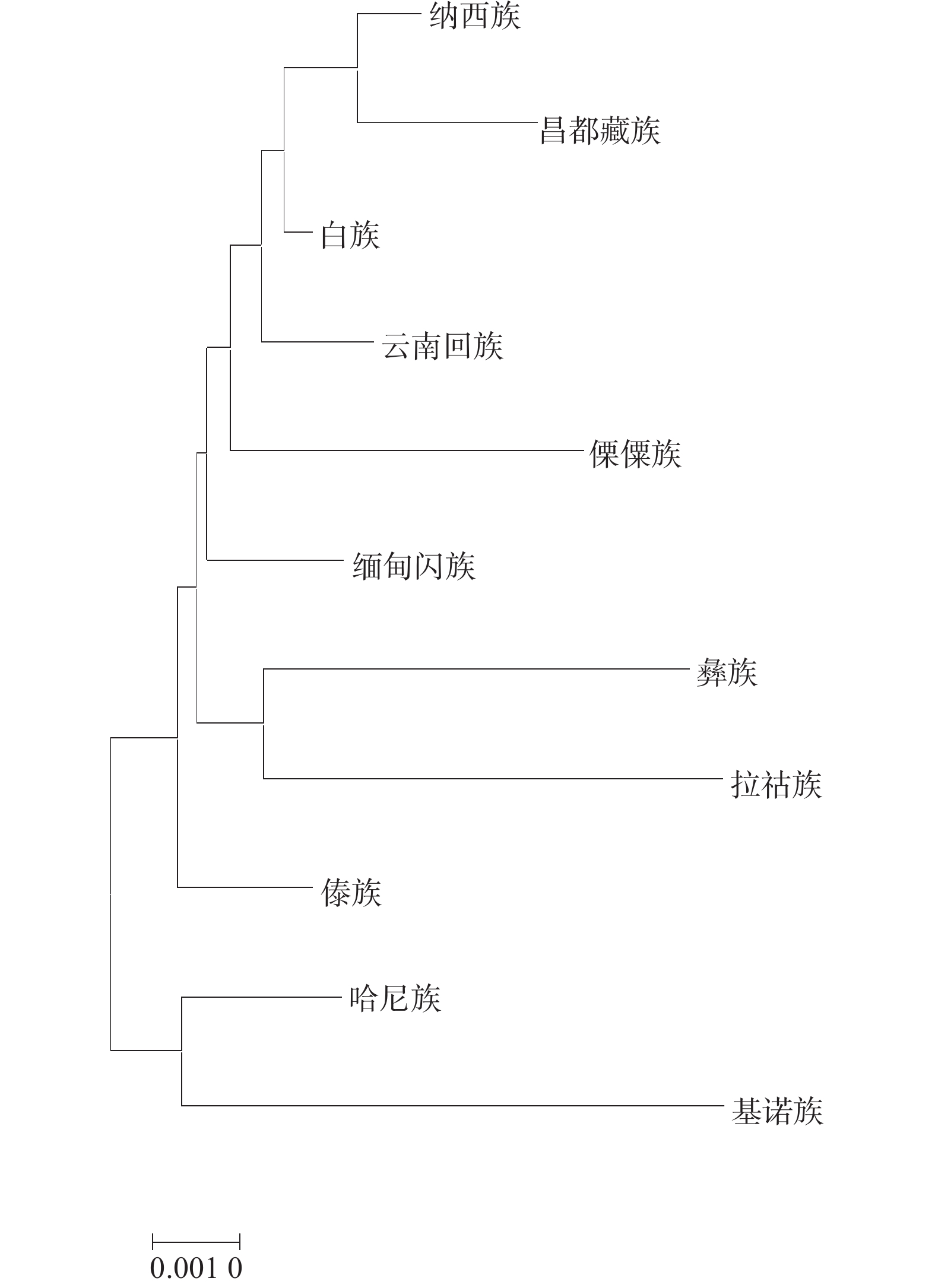
基因座 DP PIC PE Ho He HWE D3S1358 0.868776 0.669138 0.483026 0.73414 0.72136 0.87523 D13S317 0.879839 0.678767 0.453464 0.71601 0.71949 0.6294 D7S820 0.924745 0.758885 0.572409 0.7855 0.79049 0.83883 D16S539 0.89771 0.715538 0.561484 0.77946 0.757 0.56546 Penta E 0.96894 0.855748 0.680921 0.8429 0.8685 0.63086 TPOX 0.770548 0.529904 0.307136 0.61329 0.59253 0.94735 TH01 0.855432 0.637447 0.393199 0.67674 0.68133 0.72707 D2S1338 0.962496 0.84137 0.722734 0.86405 0.85763 0.33344 CSF1PO 0.865271 0.659742 0.429664 0.70091 0.71383 0.63651 D19S43 0.926552 0.775153 0.617105 0.80967 0.80222 0.138 VWA 0.915417 0.737136 0.534636 0.76435 0.77163 0.64217 D5S818 0.897874 0.711007 0.550665 0.77341 0.7489 0.42225 FGA 0.96549 0.850215 0.746972 0.87613 0.86614 0.98875 D6S1043 0.956234 0.830636 0.628515 0.81571 0.84903 0.0044 D8S1179 0.950356 0.815199 0.651601 0.82779 0.83764 0.60896 D21S11 0.939677 0.786512 0.588986 0.79456 0.8116 0.62842 D18S51 0.958206 0.834392 0.680921 0.8429 0.85193 0.00282 表 3 基诺族人群与白族、傣族、纳西族等10个人群 Standard genetic distances(左下)和Standard error(右上)
Table 3. Standard genetic distances (bottom left) and Standard error (top right) of Jino people and 10 other groups
云南回族[4] 傣族[5] 哈尼族[6] 彝族[7] 纳西族[8] 基诺族 白族[9] 缅甸闪族[10] 拉祜族[11] 昌都藏族[12] 傈僳族[13] 云南回族 − 0.0007 0.0007 0.0014 0.0003 0.0021 0.0002 0.0002 0.0017 0.0007 0.0012 傣族 0.0018 − 0.0007 0.0012 0.001 0.0014 0.0006 0.0003 0.0013 0.0014 0.0013 哈尼族 0.0017 0.0017 − 0.0013 0.0007 0.0019 0.0004 0.0005 0.0019 0.001 0.0015 彝族 0.0018 0.0016 0.0026 − 0.0007 0.0021 0.0008 0.0013 0.0017 0.001 0.0011 纳西族 0.0005 0.0025 0.0018 0.001 − 0.0021 0.0002 0.0007 0.002 0.0005 0.0014 基诺族 0.0048 0.0039 0.004 0.0049 0.0052 − 0.0022 0.0018 0.0035 0.0034 0.0032 白族 0.0001 0.0015 0.0012 0.0013 0.0004 0.0051 − 0.0002 0.0017 0.0005 0.0009 缅甸闪族 0.0001 − 0.0002 0.0006 0.0008 0.0011 0.0036 − 0.0001 − 0.0013 0.0006 0.0008 拉祜族 0.0045 0.0035 0.0052 0.0037 0.0052 0.0088 0.0044 0.0034 − 0.0028 0.0024 昌都藏族 0.0019 0.0036 0.0027 0.0025 0.0013 0.008 0.0013 0.0014 0.0071 − 0.0016 傈僳族 0.0032 0.0034 0.0044 0.0025 0.0037 0.0079 0.0026 0.0019 0.0067 0.0038 − -
[1] 国家民委网站. 基诺族[EB/OL]. (2006-4-17)[2023-4-2]. http://www.gov.cn/test/2006-04/17/content_255841.htm. [2] 赵方,伍新尧,蔡贵庆,等. Modified-Powerstates软件在法医生物统计中应用[J]. 中国法医学杂志,2003,(5):297-298,312. doi: 10.3969/j.issn.1001-5728.2003.05.015 [3] Excoffier L,Lischer H E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows[J]. Mol Ecol Resour,2010,10(3):564-567. doi: 10.1111/j.1755-0998.2010.02847.x [4] 张幼芳,张巍,侯思如. 云南回族人群17个常染色体STR基因座的遗传多态性[J]. 法医学杂志,2021,37(4):574-577. [5] Shen Z,Duan L,Yang H,et al. Genetic variation of 17 STR loci in Dai population in China's mainland[J]. Forensic Sci Int Genet,2015,19:37-38.Shen Z,Duan L,Yang H,et al. Genetic variation of 17 STR loci in Dai population in China's mainland[J]. Forensic Sci Int Genet,2015,19:37-38. [6] Huang Y,Yao J,Li J,et al. Population genetic data for 17 autosomal STR markers in the Hani population from China[J]. Int J Legal Med,2015,129(5):995-996. doi: 10.1007/s00414-015-1176-4 [7] 刘程静,郑和成,许建明,等. 云南地区彝族人群17个STR基因座的遗传多态性[J]. 中国法医学杂志,2016,31(5):493-494. doi: 10.13618/j.issn.1001-5728.2016.05.018 [8] He Y,Chen X,Duan Y,et al. Allele frequencies for fifteen autosomal STR loci in a Nakhi population from Yunnan Province,Southwest China[J]. Forensic Sci Int Genet,2016,21:e13-e14. [9] Li Y,Hong Y,Li X,et al. Allele frequency of 19 autosomal STR loci in the Bai population from the southwestern region of China's mainland[J]. Electrophoresis,2015,36(19):2498-2503. doi: 10.1002/elps.201500129Li Y,Hong Y,Li X,et al. Allele frequency of 19 autosomal STR loci in the Bai population from the southwestern region of China's mainland[J]. Electrophoresis,2015,36(19):2498-2503. doi: 10.1002/elps.201500129 [10] Chen L,Duan L,Yuan L,et al. Genetic polymorphisms of 19 autosomal STR loci in the China Burmese immigrants[J]. Forensic Sci Int Genet,2017,31:e46-e47. [11] Guo F. Genetic polymorphism of 17 autosomal STR loci in the Lahu ethnic minority from Yunnan Province,Southwest China[J]. Forensic Sci Int Genet,2017,31:e52-e53. [12] Li Z,Zhang J,Zhang H,et al. Genetic polymorphisms in 18 autosomal STR loci in the Tibetan population living in Xizang Chamdo,Southwest China[J]. Int J Legal Med,2018,132(3):733-734. doi: 10.1007/s00414-017-1740-1Li Z,Zhang J,Zhang H,et al. Genetic polymorphisms in 18 autosomal STR loci in the Tibetan population living in Xizang Chamdo,Southwest China[J]. Int J Legal Med,2018,132(3):733-734. doi: 10.1007/s00414-017-1740-1 [13] Huang Y,Zhang Y,Wang W,et al. Genetic variation of 15 STR loci in Lisu ethnic minority in southwestern China[J]. Int J Legal Med,2020,134(2):509-510. doi: 10.1007/s00414-019-02013-4 [14] Tamura K,Stecher G,Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11[J]. Mol Biol Evol,2021,38(7):3022-3027. doi: 10.1093/molbev/msab120 [15] 侯一平. 法医物证学[M]. 第4版. 北京: 人民卫生出版社, 2016: 33-34. [16] 范瑶,蒋丽霞,严江伟,等. 生物学祖孙关系鉴定的研究进展[J]. 中国法医学杂志,2023,38(01):57-62. doi: 10.13618/j.issn.1001-5728.2023.01.011 -






 下载:
下载: