Advances in Epigenetic Regulatory Mechanisms in HSV1 Infection
-
摘要: 1型单纯疱疹病毒(HSV1)是一类最为常见的人类传染病原体,感染后可导致一系列程度不同的疾病。HSV1在中枢神经系统的潜伏感染及偶发重激活是其病理发生的关键,也为抗病毒治疗带来了巨大的挑战。目前,关于HSV1感染的建立、维持和重激活的机制并未完全阐明,但普遍认为表观遗传调控可能在其中扮演重要作用。越来越多研究表明,病毒裂解期和潜伏感染期的基因组呈现不同的染色质结构,其富含的多种翻译后修饰组蛋白赋予病毒基因转录激活或抑制特征。此外,病毒潜伏相关转录本LATs也可能参与基因组表观遗传修饰调控。Abstract: Herpes simplex viruses type 1(HSV1) is among the most ubiquitous human pathogens that cause a wide variety of disease states. The latent infection of the central nervous system and sporadically reactivation is the central part of HSV1 pathogenesis, which also brings challenges to antiviral therapies. At present, the mechanism of establishing, maintaining and reactivation of HSV1 has not been fully clarified, whereas it has been generally accepted that the epigenetic regulation may play an important role. Accumulating researches have also indicated that the lytic and latent viral genomes exhibit the different chromatin structures, and the accumulation of diverse post-translational modifies the histones endow viral genes with transcriptional activation or repression features. In addition, the latency-associate transcripts of virus may also participate in the genome epigenetic modification. In this review, we summarize the research progress of epigenetic regulation of HSV1 and highlight the critical role of chromatin remodeling in HSV1 lytic proliferation and establishment of latent infection.
-
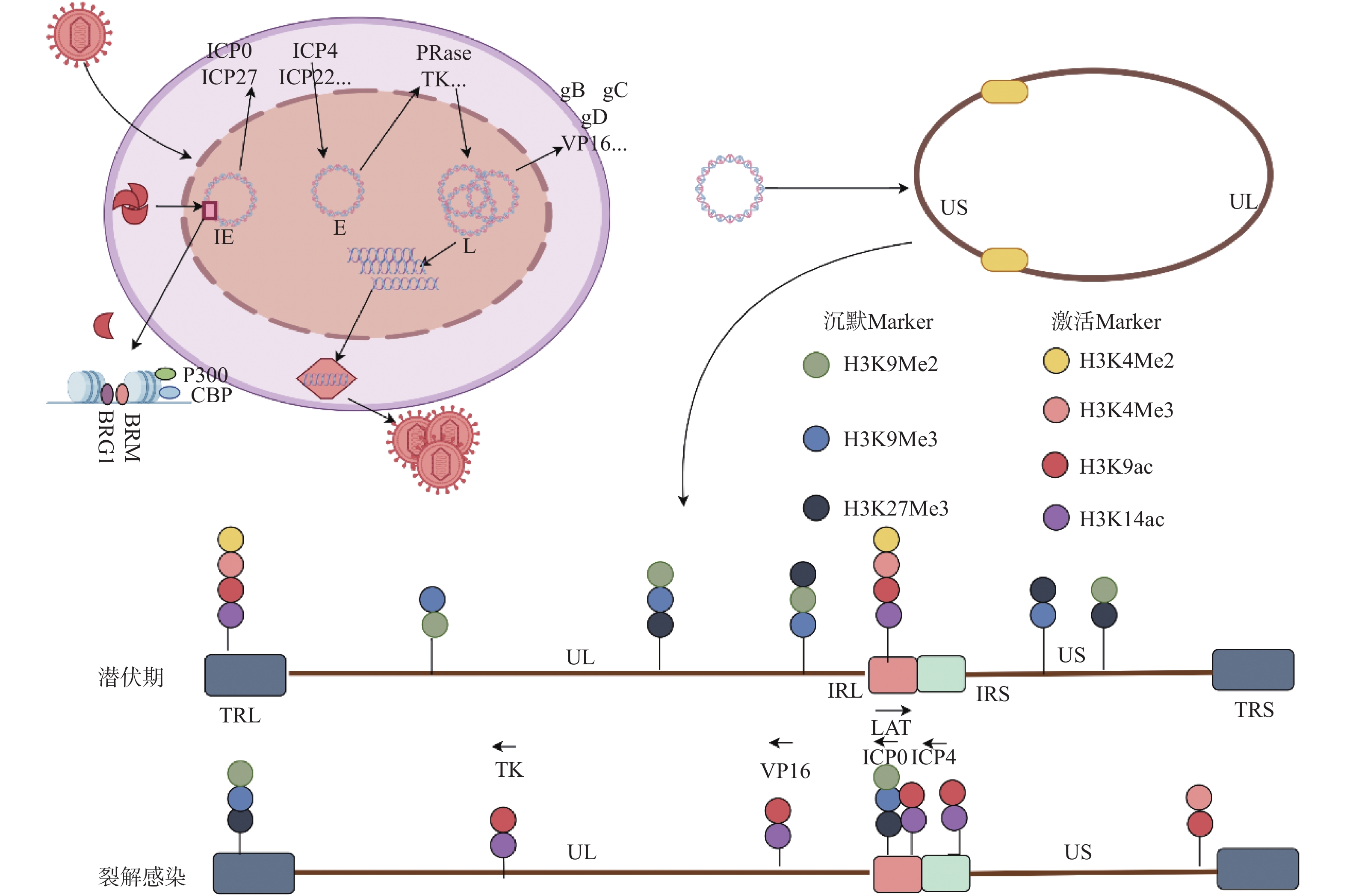
图 1 HSV1基因的级联表达和表观修饰
VP16:病毒蛋白16;IE:即早期基因;E:早期基因;L:晚期基因UL:基因组长片段区;US:基因组短片段区;RL:长片段重复区;RS:短片段重复区;图片来源于Figdraw[2] 。
Figure 1. Cascade expression and epigenetic modification of HSV1 gene
图 2 LAT上CTCF结合位点和LAT的表观调控
CTCF结合位点,包括CTRL1,CTRL2,CTa’ m,CTRS1,CTRS2,CTRS3与CTUS1[42]
Figure 2. CTCF binding sites and epigenetic regulation of LAT on LAT
-
[1] Arduino P G,Porter S R. Herpes simplex virus type 1 infection: Overview on relevant clinico-pathological features[J]. Journal of Oral Pathology & Medicine:Official Publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology,2008,37(2):107-121. [2] Schang L M,Hu M,Cortes E F,et al. Chromatin-mediated epigenetic regulation of HSV-1 transcription as a potential target in antiviral therapy[J]. Antiviral Research,2021,192(1):105103-105139. [3] Steiner I,Benninger F. Update on herpes virus infections of the nervous system[J]. Current Neurology and Neuroscience Reports,2013,13(12):414-421. doi: 10.1007/s11910-013-0414-8 [4] Lehman I R,Boehmer P E. Replication of herpes simplex virus DNA[J]. The Journal of Biological Chemistry,1999,274(40):28059-28062. doi: 10.1074/jbc.274.40.28059 [5] Knipe D M,Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection[J]. Nature Reviews Microbiology,2008,6(3):211-221. doi: 10.1038/nrmicro1794 [6] Zhu S,Viejo-Borbolla A. Pathogenesis and virulence of herpes simplex virus[J]. Virulence,2021,12(1):2670-2702. doi: 10.1080/21505594.2021.1982373 [7] Cliffe A R,Garber D A,Knipe D M. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters[J]. J Virol,2009,83(16):8182-8190. doi: 10.1128/JVI.00712-09 [8] Stevens J G,Wagner E K,Devi-rao G B,et al. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons[J]. Science (New York,NY),1987,235(4792):1056-1059. [9] Zwaagstra J C,Ghiasi H,Slanina S M,et al. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: Evidence for neuron specificity and for a large LAT transcript[J]. J Virol,1990,64(10):5019-5028. doi: 10.1128/jvi.64.10.5019-5028.1990 [10] Farrell M J,Dobson A T,Feldman L T. Herpes simplex virus latency-associated transcript is a stable intron[J]. Proceedings of the National Academy of Sciences of the United States of America,1991,88(3):790-794. [11] Pan D,Flores O,Umbach J L,et al. A neuron-specific host microRNA targets herpes simplex virus-1 ICP0 expression and promotes latency[J]. Cell Host & Microbe,2014,15(4):446-456. [12] Umbach J L,Nagel M A,Cohrs R J,et al. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia[J]. J Virol,2009,83(20):10677-10683. doi: 10.1128/JVI.01185-09 [13] Umbach J L,Kramer M F,Jurak I,et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs[J]. Nature,2008,454(7205):780-783. doi: 10.1038/nature07103 [14] Jiang X,Brown D,Osorio N,et al. Increased neurovirulence and reactivation of the herpes simplex virus type 1 latency-associated transcript (LAT)-negative mutant dLAT2903 with a disrupted LAT miR-H2[J]. Journal of Neurovirology,2016,22(1):38-49. doi: 10.1007/s13365-015-0362-y [15] Perng G C,Jones C,Ciacci-Zanella J,et al. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript[J]. Science (New York,NY),2000,287(5457):1500-1503. [16] Javier R T,Stevens J G,Dissette V B,et al. A herpes simplex virus transcript abundant in latently infected neurons is dispensable for establishment of the latent state[J]. Virology,1988,166(1):254-257. doi: 10.1016/0042-6822(88)90169-9 [17] Leib D A,Bogard C L,Kosz-Vnenchak M,et al. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency[J]. J Virol,1989,63(7):2893-2900. doi: 10.1128/jvi.63.7.2893-2900.1989 [18] Vanni E A H,Foley J W,Davison A J,et al. The latency-associated transcript locus of herpes simplex virus 1 is a virulence determinant in human skin[J]. PLoS Pathog,2020,16(12):e1009166-e1009196. doi: 10.1371/journal.ppat.1009166 [19] Moore L D,Le T,Fan G. DNA methylation and its basic function[J]. Neuropsychopharmacology:Official Publication of the American College of Neuropsychopharmacology,2013,38(1):23-38. doi: 10.1038/npp.2012.112 [20] Kouzarides T. Chromatin modifications and their function[J]. Cell,2007,128(4):693-705. doi: 10.1016/j.cell.2007.02.005 [21] Leinbach S S, Summers W C. The structure of herpes simplex virus type 1 DNA as probed by micrococcal nuclease digestion [J]. The Journal of General Virology, 1980, 51(Pt 1): 45-59. [22] Deshmane S L,Fraser N W. During latency,herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure[J]. J Virol,1989,63(2):943-947. doi: 10.1128/jvi.63.2.943-947.1989 [23] Muggeridge M I,Fraser N W. Chromosomal organization of the herpes simplex virus genome during acute infection of the mouse central nervous system[J]. J Virol,1986,59(3):764-767. doi: 10.1128/jvi.59.3.764-767.1986 [24] Huang J,Kent J R,Placek B,et al. Trimethylation of histone H3 lysine 4 by set1 in the lytic infection of human herpes simplex virus 1[J]. J Virol,2006,80(12):5740-5746. doi: 10.1128/JVI.00169-06 [25] Kent J R,Zeng P Y,Atanasiu D,et al. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription[J]. J Virol,2004,78(18):10178-10186. doi: 10.1128/JVI.78.18.10178-10186.2004 [26] Paulus C,Nitzszhe A,Nevels M. Chromatinisation of herpesvirus genomes[J]. Reviews in Medical Virology,2010,20(1):34-50. doi: 10.1002/rmv.632 [27] Lee J S,Raja P,Knipe D M. Herpesviral ICP0 protein promotes wwo waves of heterochromatin removal on an early viral promoter during lytic infection[J]. mBio,2016,7(1):e02007-e02015. [28] Gao C,Chen L,Tang S B,et al. The epigenetic landscapes of histone modifications on HSV-1 genome in human THP-1 cells[J]. Antiviral Research,2020,176(1):104730-104741. doi: 10.1016/j.antiviral.2020.104730 [29] Kubat N J,Tran R K,Mcanany P,et al. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression[J]. J Virol,2004,78(3):1139-1149. doi: 10.1128/JVI.78.3.1139-1149.2004 [30] Oh J,Fraser N W. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection[J]. J Virol,2008,82(7):3530-3537. doi: 10.1128/JVI.00586-07 [31] Gross S,Catez F,Masumoto H,et al. Centromere architecture breakdown induced by the viral E3 ubiquitin ligase ICP0 protein of herpes simplex virus type 1[J]. PloS One,2012,7(9):e44227-e44240. doi: 10.1371/journal.pone.0044227 [32] Wagner L M,Deluca N A. Temporal association of herpes simplex virus ICP4 with cellular complexes functioning at multiple steps in PolII transcription[J]. PloS One,2013,8(10):e78242-e78254. doi: 10.1371/journal.pone.0078242 [33] Herrera F J,Triezenberg S J. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection[J]. J Virol,2004,78(18):9689-9696. doi: 10.1128/JVI.78.18.9689-9696.2004 [34] Cliffe A R,Knipe D M. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection[J]. J Virol,2008,82(24):12030-12038. doi: 10.1128/JVI.01575-08 [35] Arbuckle J H,Vogel J L,Efstathiou S,et al. Deletion of the transcriptional coactivator HCF-1 in vivo impairs the removal of repressive heterochromatin from latent HSV genomes and suppresses the initiation of viral reactivation[J]. mBio,2023,14(1):e0354222-e0354238. doi: 10.1128/mbio.03542-22 [36] Bloom D C,Giordani N V,Kwiakkowski D L. Epigenetic regulation of latent HSV-1 gene expression[J]. Biochim Biophys Acta,2010,1799(3-4):246-256. doi: 10.1016/j.bbagrm.2009.12.001 [37] Neumann D M,Bhattacharjee P S,GIORDANI N V,et al. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment[J]. J Virol,2007,81(23):13248-13253. doi: 10.1128/JVI.01569-07 [38] Amelio A L,Giordani N V,Kubat N J,et al. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant[J]. J Virol,2006,80(4):2063-2068. doi: 10.1128/JVI.80.4.2063-2068.2006 [39] Kubat N J,Amelio A L,Giordani N V,et al. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription[J]. J Virol,2004,78(22):12508-12518. doi: 10.1128/JVI.78.22.12508-12518.2004 [40] Kwiatkowski D L,Thompson H W,Bloom D C. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency[J]. J Virol,2009,83(16):8173-8181. doi: 10.1128/JVI.00686-09 [41] Wang Q Y,Zhou C,Johnson K E,et al. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection[J]. Proceedings of the National Academy of Sciences of the United States of America,2005,102(44):16055-16059. [42] Amelio A L,Mcanany P K,Bloom D C. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities[J]. J Virol,2006,80(5):2358-2368. doi: 10.1128/JVI.80.5.2358-2368.2006 [43] Lang F,Li X,Vladimirova O,et al. CTCF interacts with the lytic HSV-1 genome to promote viral transcription[J]. Scientific Reports,2017,7(1):39861-39876. doi: 10.1038/srep39861 [44] Washington S D,Musarrat F,ERTEL M K,et al. CTCF binding sites in the herpes simplex virus 1 genome display site-specific CTCF occupation,protein recruitment,and insulator function[J]. J Virol,2018,92(8):e00156-e00171. [45] Washington S D,Singh P,Johns R N,et al. The CCCTC binding factor,CTRL2,modulates heterochromatin deposition and the establishment of herpes shimplex virus 1 latency in vivo[J]. J Virol,2019,93(13):e00415-e00419. [46] Bedadala G R,Pinnoji R C,Palem J R,et al. Thyroid hormone controls the gene expression of HSV-1 LAT and ICP0 in neuronal cells[J]. Cell Res,2010,20(5):587-598. doi: 10.1038/cr.2010.50 [47] Grams T R,Edwards T G,Bloom D C. HSV-1 LAT promoter deletion viruses exhibit strain-specific and LAT-dependent epigenetic regulation of latent viral genomes in human neurons[J]. J Virol,2023,97(2):e01935-e01951. -






 下载:
下载: