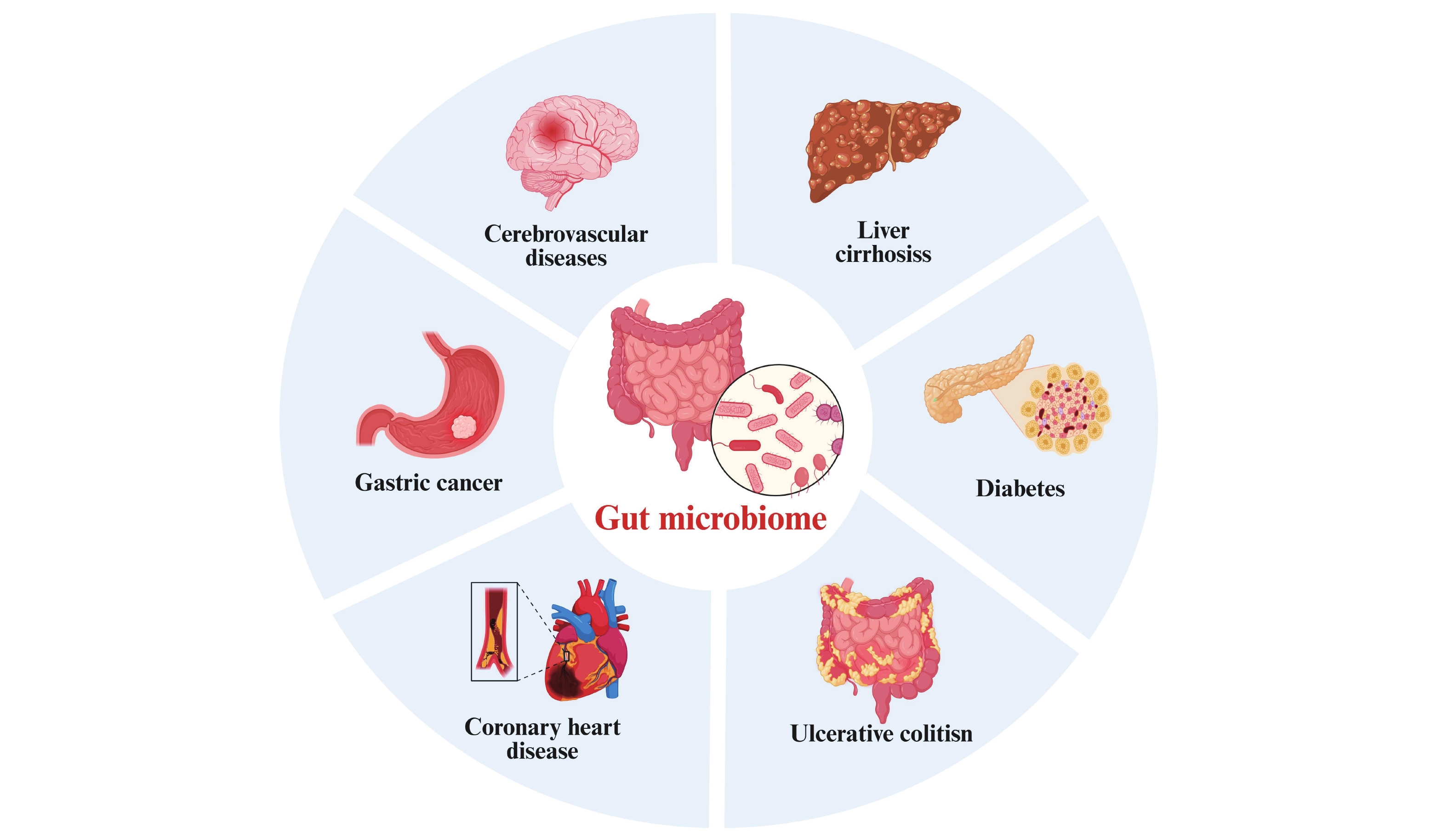
Regional and Ethnic-Specific Characteristics of the Gut Microbiome in China and Their Impact on Disease
-
摘要: 肠道微生物组(gut microbiome,GM)的构成受地理、民族、饮食和遗传等诸多因素调控。系统综述了中国不同地域与少数民族的GM研究,重点阐释地理因素和民族背景对GM组成与结构的影响,并深入探讨少数民族特异性菌群及其代谢功能与疾病易感性的关联。通过系统比较不同群体在菌群相关疾病中的微生物组成差异,旨在揭示其内在规律,以期为民族特异性的GM研究、疾病机制解析及精准防控策略的制定提供理论依据。Abstract: The composition of the Gut Microbiome (GM) is regulated by numerous factors, including geography, ethnicity, diet, and genetics. This review systematically synthesizes studies on the GM across different geographic regions and ethnic minority groups in China. It focuses on elucidating the impact of geographical factors and ethnic backgrounds on GM composition and structure and provides an in-depth discussion on the association between ethnicity-specific microbial taxa, their metabolic functions, and susceptibility to diseases. By systematically comparing microbial compositional differences in microbiota-related diseases among different populations, this review aims to uncover the underlying patterns. The findings are expected to provide a theoretical foundation for ethnicity-specific GM research, the elucidation of disease mechanisms, and the development of targeted prevention and control strategies.
-
Key words:
- Gut microbiota /
- Microbiota composition /
- Ethnic minorities /
- Regional variation
-
表 1 中国部分少数民族成员的特征性肠道微生物组
Table 1. Characteristic gut microbial signatures in selected Chinese ethnic minority populations
序号 民族 菌群分类 序号 民族 菌群分类 1 赫哲族 芽生菌属 6 白族 乳酸菌属
双歧杆菌属2 彝族 普雷沃氏菌属
拟杆菌属7 瑶族 拟杆菌属 3 黎族 巨球型菌属 8 藏族 粪杆菌属
Psammococcus4 壮族 戈氏副拟杆菌
动物双歧杆菌9 蒙古族 普雷沃氏菌属 5 哈尼族 汉逊德巴利酵母菌
禾谷镰刀菌10 维吾尔族
哈萨克族瘤状球菌科
链球菌科
肠杆菌科表 2 中国不同民族人群肠道菌群与疾病关联研究
Table 2. Studies on the association between gut microbiota and diseases in different ethnic groups in china
序号 民族 疾病 肠道菌群 结论 1 维吾尔族
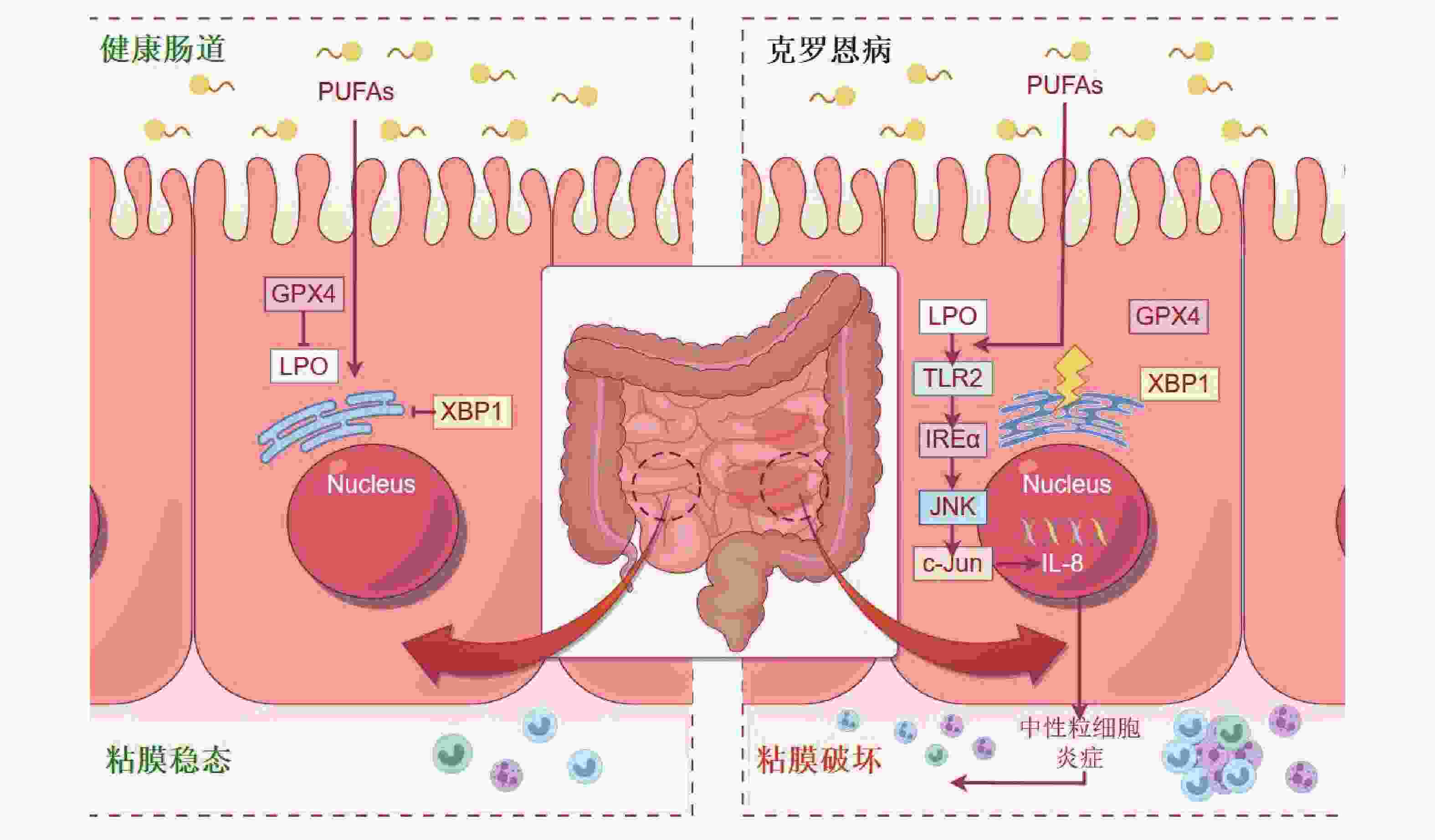
汉族溃疡性结肠炎 伯克霍尔德菌属
帕拉伯克霍尔普拉菌属肠道微生物群落的组成差异可能是导致维吾尔族和汉族患者溃疡性结肠炎(UC)临床表现差异的一个因素。嗜血杆菌可能成为预测 UC 的潜在生物标志物[56]。 2 藏族
汉族肠易激综合征 厚壁菌门、拟杆菌门
布劳特氏菌属由于藏族和汉族肠易激综合征患者肠道微生物群特征存在差异,布劳特氏菌可能会使患有肠易激综合征的藏族患者更容易出现腹胀症状[59]。 3 藏族 肝硬化 厌氧杆菌属
双歧杆菌属
多尔氏菌属藏族肝硬化患者体内拟杆菌属和放线菌属的数量显著增多[63]。 4 土家族
苗族
汉族缺血性脑卒中 肠球菌科
链球菌科传统的信息系统风险因素(收缩压、舒张压以及高敏C反应蛋白)与少数民族宿主的肠道微生物群之间存在相互作用[66]。 5 藏族 冠状动脉疾病 卡特尼巴克斯特菌
霍尔德曼纳菌
瘤胃球菌高海拔地区藏族人的独特生活环境和饮食习惯调节了其肠道微生物群的变化,从而使得他们患冠状动脉疾病(CAD)的几率低于普通人群[67]。 6 藏族 冠状动脉
心脏病小杆菌属、苏卡尼维菌属
布劳特氏菌属民族背景和环境对藏族人的肠道微生物群有着显著影响。冠心病(CHD)与藏族人的肠道微生物群密切相关[68]。 7 傣族
汉族II型糖尿病 拟杆菌属
变形菌门在汉族人群中,拟杆菌属数量的显著增加与II型糖尿病(T2DM)的发生有关;而在傣族人群中,变形菌属数量的显著增加则与T2DM的发生有关[72]。 8 哈萨克族
维吾尔族
蒙古族糖尿病 梭状芽孢杆菌
气单胞菌科
脱硫弧菌科
埃瑞斯皮洛特菌科不同种族群体中与糖尿病相关的细菌在组成和功能上存在显著差异。在研究糖尿病与肠道微生物群之间的关联时,应充分考虑种族差异的影响[75-76]。 -
[1] Tan G S E, Tay H L, Tan S H, et al. Gut microbiota modulation: Implications for infection control and antimicrobial stewardship[J]. Adv Ther, 2020, 37(10): 4054-4067. doi: 10.1007/s12325-020-01458-z [2] Trueba G, Cardenas P, Romo G, et al. Reevaluating human-microbiota symbiosis: Strain-level insights and evolutionary perspectives across animal species[J]. Biosystems, 2024, 244: 105283. doi: 10.1016/j.biosystems.2024.105283 [3] Castagnoli R, Pala F, Bosticardo M, et al. Gut microbiota-host interactions in inborn errors of immunity[J]. Int J Mol Sci, 2021, 22(3): 1416. doi: 10.3390/ijms22031416 [4] Mamun M A A, Rakib A, Mandal M, et al. Impact of a high-fat diet on the gut microbiome: A comprehensive study of microbial and metabolite shifts during obesity[J]. Cells, 2025, 14(6): 463. doi: 10.3390/cells14060463 [5] Liu M, Nieuwdorp M, de Vos W M, et al. Microbial tryptophan metabolism tunes host immunity, metabolism, and extraintestinal disorders[J]. Metabolites, 2022, 12(9): 834. doi: 10.3390/metabo12090834 [6] Ahrend H, Buchholtz A, Stope M B. Microbiome and mucosal immunity in the intestinal tract[J]. In Vivo, 2025, 39(1): 17-24. doi: 10.21873/invivo.13801 [7] Howard A, Carroll-Portillo A, Alcock J, et al. Dietary effects on the gut phageome[J]. Int J Mol Sci, 2024, 25(16): 8690. doi: 10.3390/ijms25168690 [8] Haddad E N, Sugino K Y, Tucker R M, et al. Gut enterotypes are stable during Bifidobacterium and Lactobacillus probiotic supplementation[J]. Journal of Food Science, 2020, 85(5): 1596-1604. doi: 10.1111/1750-3841.15127 [9] Russell B J, Verma M, Maier N K, et al. Dissecting host-microbe interactions with modern functional genomics[J]. Curr Opin Microbiol, 2024, 82: 102554. doi: 10.1016/j.mib.2024.102554 [10] Kwok LY, Zhang J, Guo Z, et al. Characterization of fecal microbiota across seven Chinese ethnic groups by quantitative polymerase chain reaction[J]. PLoS One, 2014, 9(4): e93631. doi: 10.1371/journal.pone.0093631 [11] Pasolli E, Asnicar F, Manara S, et al. Extensive unexplored human microbiome diversity revealed by over 150, 000 genomes from metagenomes spanning age, geography, and lifestyle[J]. Cell, 2019, 176(3): 649-662. e20. [12] Yatsunenko T, Rey F E, Manary M J, et al. Human gut microbiome viewed across age and geography[J]. Nature, 2012, 486(7402): 222-227. doi: 10.1038/nature11053 [13] Gaulke C A, Sharpton T J. The influence of ethnicity and geography on human gut microbiome composition[J]. Nat Med, 2018, 24(10): 1495-1496. doi: 10.1038/s41591-018-0210-8 [14] Nakayama J, Watanabe K, Jiang J, et al. Diversity in gut bacterial community of school-age children in Asia[J]. Sci Rep, 2015, 5: 8397. doi: 10.1038/srep08397 [15] He Y, Wu W, Zheng H M, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models[J]. Nat Med, 2018, 24(10): 1532-1535. doi: 10.1038/s41591-018-0164-x [16] Han N, Zhang T, Qiang Y, et al. Time-scale analysis of the long-term variability of human gut microbiota characteristics in Chinese individuals[J]. Commun Biol, 2022, 5(1): 1414. doi: 10.1038/s42003-022-04359-9 [17] Lu J, Zhang L, Zhai Q, et al. Chinese gut microbiota and its associations with staple food type, ethnicity, and urbanization[J]. NPJ Biofilms Microbiomes, 2021, 7(1): 71. doi: 10.1038/s41522-021-00245-0 [18] Zhang J, Qi H, Li M, et al. Diet mediate the impact of host habitat on gut microbiome and influence clinical indexes by modulating gut microbes and serum metabolites[J]. Adv Sci (Weinh), 2024, 11(19): e2310068. doi: 10.1002/advs.202310068 [19] Zhang J, Guo Z, Xue Z, et al. A Phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities[J]. ISME J, 2015, 9(9): 1979-1990. doi: 10.1038/ismej.2015.11 [20] Zhang X, Zhong H, Li Y, et al. Sex- and age-related trajectories of the adult human gut microbiota shared across populations of different ethnicities[J]. Nat Aging, 2021, 1(1): 87-100. doi: 10.1038/s43587-020-00014-2 [21] Sun Y, Zuo T, Cheung C P, et al. Population-level configurations of gut mycobiome across 6 ethnicities in urban and rural China[J]. Gastroenterology, 2021, 160(1): 272-286. e11. [22] 戚宇锋, 任来峰, 曹雪玲, 等. 山西省健康人群的肠道菌群组成特征及其与性别和年龄的关系[J]. 中国微生态学杂志, 2019, 31(10): 1117-1123. [23] Yu Y, Yang X, Liu D, et al. Ancient genomic analysis of a Chinese hereditary elite from the Northern and Southern Dynasties[J]. J Genet Genom, 2025, 52(4): 473-482. doi: 10.1016/j.jgg.2024.07.009 [24] Chen R, Duan Z Y, Duan X H, et al. Progress in research on gut microbiota in ethnic minorities in China and consideration of intervention strategies based on ethnic medicine: A review[J]. Front Cell Infect Microbiol, 2022, 12: 1027541. doi: 10.3389/fcimb.2022.1027541 [25] Bosch J A, Nieuwdorp M, Zwinderman A H, et al. The gut microbiota and depressive symptoms across ethnic groups[J]. Nat Commun, 2022, 13(1): 7129. doi: 10.1038/s41467-022-34504-1 [26] Keohane D M, Ghosh T S, Jeffery I B, et al. Microbiome and health implications for ethnic minorities after enforced lifestyle changes[J]. Nat Med, 2020, 26(7): 1089-1095. doi: 10.1038/s41591-020-0963-8 [27] 黄卫强, 张家超, 郑艺, 等. 白族成年人肠道菌群多样性研究[J]. 微生物学通报, 2015, 42(3): 504-515. [28] Xiao Y, Zhai Q, Zhang H, et al. Gut colonization mechanisms of Lactobacillus and Bifidobacterium: An argument for personalized designs[J]. Annu Rev Food Sci Technol, 2021, 12: 213-233. doi: 10.1146/annurev-food-061120-014739 [29] Li S, Li Y, Wang W, et al. Dietary habits of pastoralists on the Tibetan Plateau are influenced by remoteness and economic status[J]. Food Res Int, 2023, 174: 113627. doi: 10.1016/j.foodres.2023.113627 [30] Lawrence E S, Gu W, Bohlender R J, et al. Functional EPAS1/HIF2A missense variant is associated with hematocrit in Andean highlanders[J]. Sci Adv, 2024, 10(6): eadj5661. doi: 10.1126/sciadv.adj5661 [31] Yang Z, Gao J, Sun J, et al. Gut microbiota in Tibetan, Mongolian and Zhuang children aged 10-12 years old in 2016[J]. Wei Sheng Yan Jiu, 2022, 51(3): 411-416. [32] Liu W, Zhang J, Wu C, et al. Unique Features of Ethnic Mongolian Gut Microbiome revealed by metagenomic analysis[J]. Sci Rep, 2016, 6: 34826. doi: 10.1038/srep34826 [33] Li J, Xu H, Sun Z, et al. Effect of dietary interventions on the intestinal microbiota of Mongolian hosts[J]. Sci Bull, 2016, 61(20): 1605-1614. doi: 10.1007/s11434-016-1173-0 [34] Zhang J, Guo Z, Lim A A, et al. Mongolians core gut microbiota and its correlation with seasonal dietary changes[J]. Sci Rep, 2014, 4: 5001. doi: 10.1038/srep05001 [35] Liao M, Xie Y, Mao Y, et al. Comparative analyses of fecal microbiota in Chinese isolated Yao population, minority Zhuang and rural Han by 16sRNA sequencing[J]. Sci Rep, 2018, 8(1): 1142. doi: 10.1038/s41598-017-17851-8 [36] Finley J W, Burrell J B, Reeves P G. Pinto bean consumption changes SCFA profiles in fecal fermentations, bacterial populations of the lower bowel, and lipid profiles in blood of humans[J]. J Nutr, 2007, 137(11): 2391-2398. doi: 10.1093/jn/137.11.2391 [37] Tang W H, Kitai T, Hazen S L. Gut microbiota in cardiovascular health and disease[J]. Circ Res, 2017, 120(7): 1183-1196. doi: 10.1161/CIRCRESAHA.117.309715 [38] 彭倩楠, 霍冬雪, 徐传标, 等. 黎族人肠道微生物群落结构特征及其与饮食关联性[J]. 微生物学通报, 2017, 44(11): 2624-2633. [39] 兰菊杰, 王春敏, 崔国利, 等. 赫哲族和汉族健康人群肠道细菌群落分析[J]. 中国微生态学杂志, 2021, 33(5): 513-518. [40] Palmieri L J, Rainteau D, Sokol H, et al. Inhibitory effect of ursodeoxycholic acid on Clostridium difficile germination is insufficient to prevent colitis: A study in hamsters and humans[J]. Front Microbiol, 2018, 9: 2849. doi: 10.3389/fmicb.2018.02849 [41] 杨倬, 孙静, 高洁, 等. 维吾尔族与汉族儿童肠道细菌群落分析[J]. 微生物学通报, 2020, 47(11): 3833-3842. [42] Wang Y, Luo X, Mao X, et al. Gut microbiome analysis of type 2 diabetic patients from the Chinese minority ethnic groups the Uygurs and Kazaks[J]. PLoS One, 2017, 12(3): e0172774. doi: 10.1371/journal.pone.0172774 [43] 郑艺, 张家超, 乔健敏, 等. 壮族人群肠道菌群多样性分析[J]. 中国食品学报, 2016, 16(1): 226-236. [44] 蒋康, 宿仁琴. 石棉地区藏族、彝族和汉族人群肠道微生物菌群构成与分布[J]. 中国微生态学杂志, 2020, 32(7): 777-781. [45] Bai X, Sun Y, Li Y, et al. Landscape of the gut archaeome in association with geography, ethnicity, urbanization, and diet in the Chinese population[J]. Microbiome, 2022, 10(1): 147. doi: 10.1186/s40168-022-01335-7 [46] Doran S, Arif M, Lam S, et al. Multi-omics approaches for revealing the complexity of cardiovascular disease[J]. Brief Bioinform, 2021, 22(5): bbab061. doi: 10.1093/bib/bbab061 [47] Bolte L A, Vich Vila A, Imhann F, et al. Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome[J]. Gut, 2021, 70(7): 1287-1298. doi: 10.1136/gutjnl-2020-322670 [48] Qi Q, Li J, Yu B, et al. Host and gut microbial tryptophan metabolism and type 2 diabetes: An integrative analysis of host genetics, diet, gut microbiome and circulating metabolites in cohort studies[J]. Gut, 2022, 71(6): 1095-1105. doi: 10.1136/gutjnl-2021-324053 [49] Kazemian N, Mahmoudi M, Halperin F, et al. Gut microbiota and cardiovascular disease: Opportunities and challenges[J]. Microbiome, 2020, 8(1): 36. doi: 10.1186/s40168-020-00821-0 [50] Li H, Zhang H, Hua W, et al. Causal relationship between gut microbiota and functional outcomes after ischemic stroke: A comprehensive Mendelian randomization study[J]. J Stroke Cerebrovasc Dis, 2024, 33(8): 107814. doi: 10.1016/j.jstrokecerebrovasdis.2024.107814 [51] Aho V T E, Houser M C, Pereira P A B, et al. Relationships of gut microbiota, short-chain fatty acids, inflammation, and the gut barrier in Parkinson’s disease[J]. Mol Neurodegener, 2021, 16(1): 6. doi: 10.1186/s13024-021-00427-6 [52] Zhang J, Dong C, Lin Y, et al. Causal relationship between gut microbiota and gastric cancer: A two-sample Mendelian randomization analysis[J]. Mol Clin Oncol, 2024, 20(5): 38. doi: 10.3892/mco.2024.2736 [53] Li J, Wang M, Ma S, et al. Association of gastrointestinal microbiome and obesity with gestational diabetes mellitus-an updated globally based review of the high-quality literatures[J]. Nutr Diabetes, 2024, 14(1): 31. doi: 10.1038/s41387-024-00291-5 [54] Yu R L, Weber H C. Irritable bowel syndrome, the gut microbiome, and diet[J]. Curr Opin Endocrinol Diabetes Obes, 2025, 32(3): 102-107. doi: 10.1097/MED.0000000000000905 [55] Zhou C, Xu M, Xiao Z, et al. Distribution of HLA-DQA1-DQB1 and-DRB1 genes and haplotypes in Han, Uyghur, Kazakh and Hui populations inhabiting Xinjiang Uyghur Autonomous Region, China[J]. Int J Immunogenetics, 2021, 48(3): 229-238. doi: 10.1111/iji.12529 [56] Liu H, Liu W, Huang X, et al. Intestinal flora differences between patients with ulcerative colitis of different ethnic groups in China[J]. Medicine (Baltimore), 2021, 100(32): e26932. [57] Yao P, Cui M, Wang H, et al. Quantitative analysis of intestinal flora of uygur and Han ethnic Chinese patients with ulcerative colitis[J]. Gastroenterol Res Pract, 2016, 2016: 9186232. [58] Vakili O, Adibi Sedeh P, Pourfarzam M. Metabolic biomarkers in irritable bowel syndrome diagnosis[J]. Clin Chim Acta, 2024, 560: 119753. doi: 10.1016/j.cca.2024.119753 [59] Ma X, Huan H, Liu C, et al. Influence of ethnic origin on the clinical characteristics and intestinal flora of irritable bowel syndrome: A prospective study between Han and Tibetan patients[J]. Front Med (Lausanne), 2024, 11: 1359962. doi: 10.3389/fmed.2024.1359962 [60] Liu X, Mao B, Gu J, et al. Blautia-a new functional genus with potential probiotic properties?[J]. Gut Microbes, 2021, 13(1): 1-21. [61] Yang Y, Schnabl B. Gut bacteria in alcohol-associated liver disease[J]. Clin Liver Dis, 2024, 28(4): 663-679. doi: 10.1016/j.cld.2024.06.008 [62] Assimakopoulos S F, Bhagani S, Aggeletopoulou I, et al. The role of gut barrier dysfunction in postoperative complications in liver transplantation: Pathophysiological and therapeutic considerations[J]. Infection, 2024, 52(3): 723-736. doi: 10.1007/s15010-024-02182-4 [63] Huan H, Ren T, Xu L, et al. Compositional distinction of gut microbiota between Han Chinese and Tibetan populations with liver cirrhosis[J]. PeerJ, 2021, 9: e12142. doi: 10.7717/peerj.12142 [64] Miah L, Robertson N. The gut microbiome: Neurological development and disease[J]. J Neurol, 2022, 269(10): 5682-5684. doi: 10.1007/s00415-022-11334-1 [65] Laryushina Y, Samoilova-Bedych N, Turgunova L, et al. Alterations of the gut microbiome and TMAO levels in patients with ulcerative colitis[J]. J Clin Med, 2024, 13(19): 5794. doi: 10.3390/jcm13195794 [66] Zhang N, Wang H, Wang X, et al. Combination effect between gut microbiota and traditional potentially modifiable risk factors for first-ever ischemic stroke in Tujia, Miao and Han populations in China[J]. Front Mol Neurosci, 2022, 15: 922399. doi: 10.3389/fnmol.2022.922399 [67] Ma Y, Zhu L, Ma Z, et al. Distinguishing feature of gut microbiota in Tibetan highland coronary artery disease patients and its link with diet[J]. Sci Rep, 2021, 11(1): 18486. doi: 10.1038/s41598-021-98075-9 [68] Liu F, Fan C, Zhang L, et al. Alterations of gut microbiome in Tibetan patients with coronary heart disease[J]. Front Cell Infect Microbiol, 2020, 10: 373. doi: 10.3389/fcimb.2020.00373 [69] Cheng Q, Fan C, Liu F, et al. Structural and functional dysbiosis of gut microbiota in Tibetan subjects with coronary heart disease[J]. Genomics, 2022, 114(6): 110483. doi: 10.1016/j.ygeno.2022.110483 [70] Hamjane N, Mechita M B, Nourouti N G, et al. Gut microbiota dysbiosis-associated obesity and its involvement in cardiovascular diseases and type 2 diabetes. A systematic review[J]. Microvasc Res, 2024, 151: 104601. doi: 10.1016/j.mvr.2023.104601 [71] Zhang L, Sun Y, Zhang X, et al. Three novel genetic variants in the FAM110D CACNA1A and NLRP12 genes are associated with susceptibility to hypertension among Dai people[J]. Am J Hypertens, 2021, 34(8): 874-879. doi: 10.1093/ajh/hpab040 [72] Tang L T, Feng L, Cao H Y, et al. Comparative study of type 2 diabetes mellitus-associated gut microbiota between the Dai and Han populations[J]. World J Diabetes, 2023, 14(12): 1766-1783. doi: 10.4239/wjd.v14.i12.1766 [73] Liu Y, Wang M, Li W, et al. Differences in gut microbiota and its metabolic function among different fasting plasma glucose groups in Mongolian population of China[J]. BMC Microbiol, 2023, 23(1): 102. doi: 10.1186/s12866-023-02852-7 [74] Zhang X, Yang X L, Liu S, et al. Prevalence of hypertension among type 2 diabetes mellitus patients in China: A systematic review and meta-analysis[J]. Int Health, 2024, 16(2): 144-151. doi: 10.1093/inthealth/ihad047 [75] Li S C, Xiao Y, Wu R T, et al. Comparative analysis of type 2 diabetes-associated gut microbiota between Han and Mongolian people[J]. J Microbiol, 2021, 59(7): 693-701. doi: 10.1007/s12275-021-0454-8 [76] Park S K, Jung J Y, Oh C M, et al. Components of metabolic syndrome and their relation to the risk of incident cerebral infarction[J]. Endocr J, 2021, 68(3): 253-259. doi: 10.1507/endocrj.EJ20-0486 -





 下载:
下载: