Genomic Analysis of Two Novel Coxsackievirus B2 Strains in 2022
-
摘要:
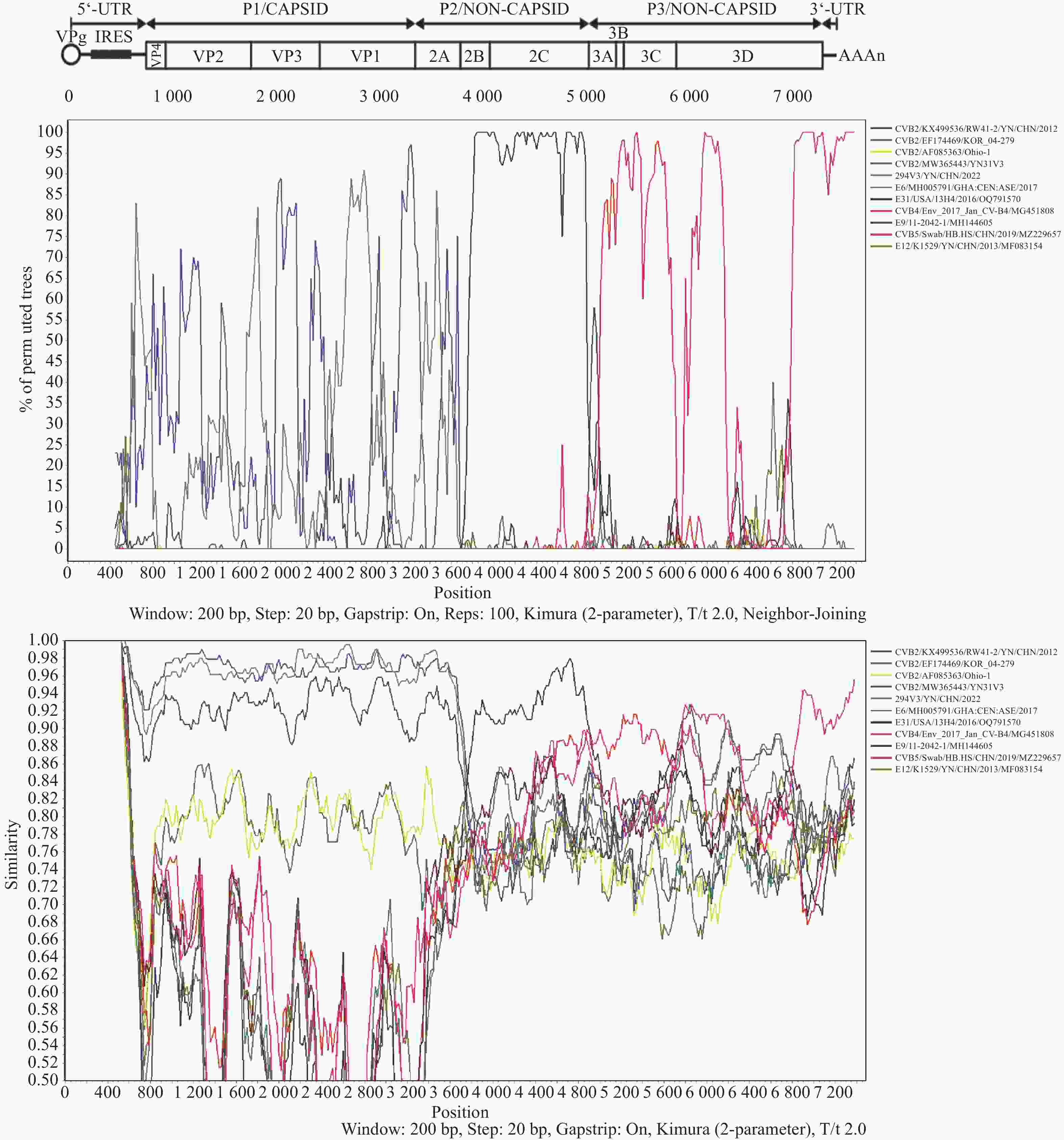
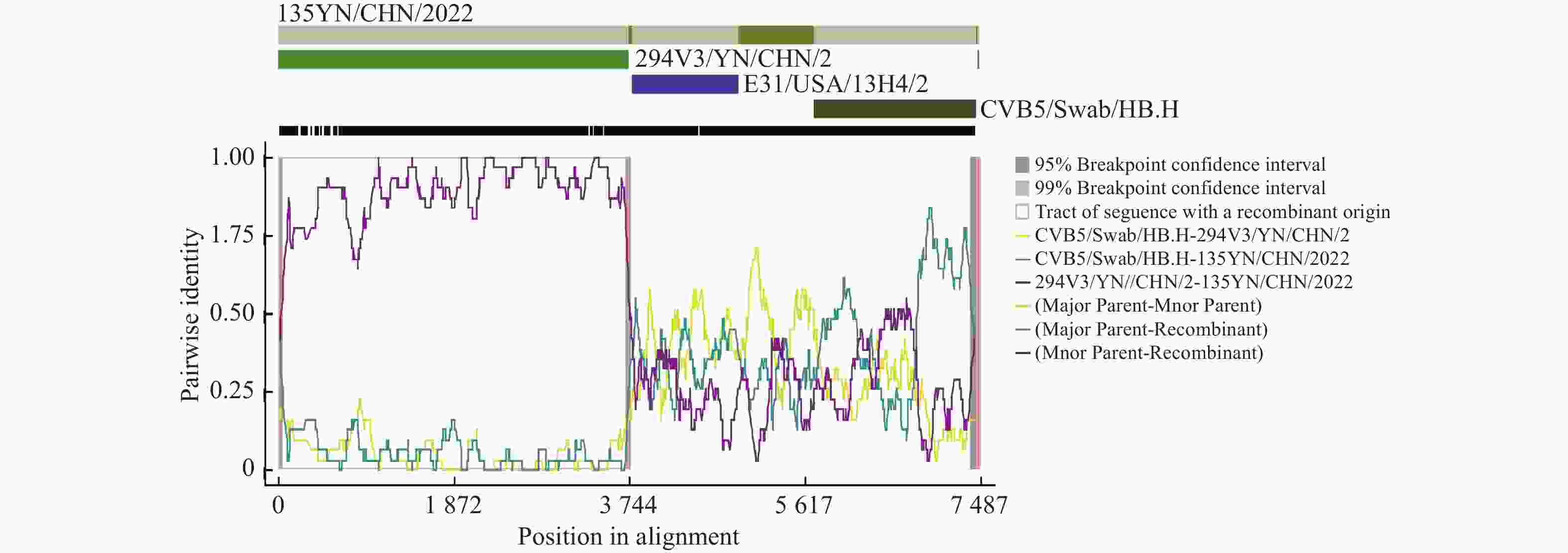
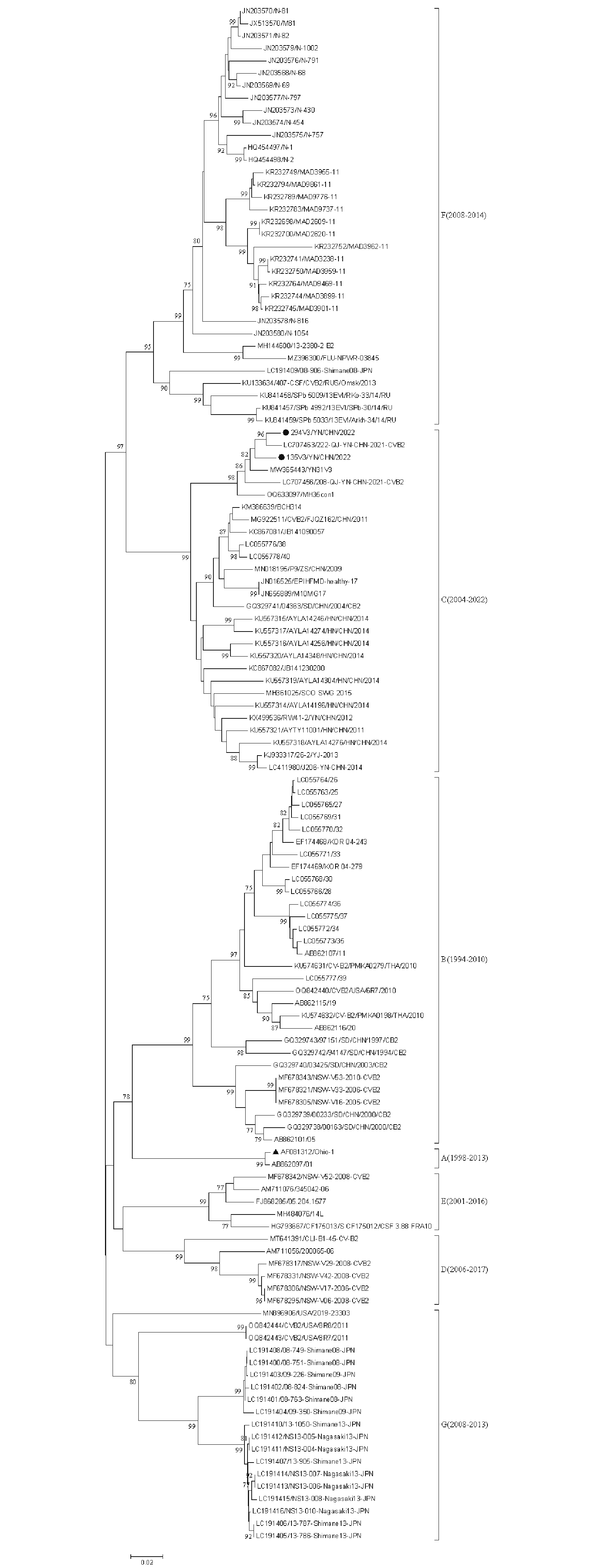
目的 对云南省柯萨奇病毒B组2型重组毒株进行全基因组分析。 方法 设计覆盖CVB2全基因组序列的扩增引物,采用分段PCR策略获取病毒基因组片段,通过"引物移步法"完成测序,随后进行序列组装。运用DNAStar 7.1、 MEGA 7.0、 Simplot 3.5.1与RDP4等软件进行全基因组序列分析。 结果 VP1区系统发育分析显示,两株CVB2分离株均属C基因型,与云南地区近五年流行株亲缘关系最近。全基因组序列分析显示,两株分离株间核苷酸一致性为90.1%。P1-P3区进化分析表明,P2和P3区域中CVB2毒株与非CVB2血清型EVB毒株聚类。通过Simplot和Bootscan分析,发现135V3/YN/CHN/2022分离株在非结构编码区可能存在多个重组事件。此外,RDP4分析结果显示135V3/YN/CHN/2022分离株经历了涉及E31/USA/13H4/2016和CVB5/Swab/HB.HS/CHN/2019分离株的重组事件。 结论 2022年云南昆明分离的两株CVB2属于C基因型并存在重组,凸显了持续监测其基因组变化的必要性 Abstract:Objective To perform whole-genome analysis of recombinant Coxsackievirus B2 (CVB2) strains isolated in Yunnan Province. Methods Primers spanning the complete CVB2 genomic sequence were designed. Viral genomic fragments were amplified using a segmented PCR strategy, followed by sequencing via primer walking and subsequent sequence assembly. Comprehensive genomic analyses were conducted using DNAStar 7.1, MEGA 7.0, Simplot 3.5.1, and RDP4 software. Results Phylogenetic analysis of the VP1 region confirmed both CVB2 isolates belonged to genotype C. Evolutionary assessment of P1-P3 regions revealed clustering of CVB2 strains with diverse EVB strains in P2 and P3 regions. Simplot and Bootscan analyses identified potential multiple recombination events within non-structural coding regions of isolate 135V3/YN/CHN/2022. Furthermore, RDP4 analysis demonstrated that isolate 135V3/YN/CHN/2022 underwent recombination events involving strains E31/USA/13H4/2016 and CVB5/Swab/HB. HS/CHN/2019. Conclusion The two CVB2 strains, isolated in Kunming Yunnan in 2022 belonged to genotype C and exhibited recombination events, highlighting the need for continuous monitoring of their genomic changes. -
Key words:
- Enterovirus /
- Coxsackievirus B2 /
- Whole genome /
- Sequence analysis /
- Recombination analysis
-
表 1 全基因扩增和测序引物
Table 1. Amplification and sequencing primers of whole genome
引物名称 引物序列(5'-3') 位置 BOAS GGTGCTCACTAGGAGGTCYCT 3505 –3484 BOS GGYTAYATNCANTGYTGGTAYCARA 2329 –2324 E201F TTAAAACAGCCTGTGGGTTG 1–20 B2N1r GCCAATCCAATAGCTATAT 586–568 B2N1f GTGCTAGTGGAAGTTCCA 795–812 B2N2r CTGTGGCCATTTACCATA 1069 -1052 B2N2R CCAATAGTGTCAGCAACTC 2511 -2493 B2N3F TAACTGATGTGACGGAAAAG 3261 –3280 B2N4f TCGCCCTCATAGGCTGCA 3977 –3960 B22944f TGGCCCTCATAGGCTGCA 3960 -3977 B2N5f CCATCCAGTTTATTGATAG 4914 -4932 B22945f GATGGTACCAACCTGGAA 5564 –5581 B2N6f GAGGAAYGGGGACCCAAG 6046 -6043 B2N6r ACCGNCAAGCATAACTGG 6598 –6581 B2N1356r AAACAAGCAAACCAYAC 6663 –6647 B2N7f GAAGAGGTTTTTCAGGGC 7042 –7059 E208R ACCGAATGCGGAGAATTTAC 7404 –7385 注:BOAS、BOS、E201F、B2N2R、B2N3F、E208R为扩增引物,其余引物为测序引物。 表 2 135V3/YN/CHN/2022分离株与原型株及其他CVB2分离株不同基因组区域的核苷酸和氨基酸序列一致性
Table 2. Nucleotide and amino acid sequence identity between 135V3/YN/CHN/2022 isolates and the prototype strain and other CVB2 isolates in all sequenced genomic regions
基因组区域 原型株 其他CVB2分离株 核苷酸一致性(%) 氨基酸一致性(%) 核苷酸一致性(%) 氨基酸一致性(%) 5’UTR 84.4 85.3-97.0 VP4 82.6 92.8 78.3-95.7 91.3-97.1 VP2 82.7 96.2 81.3-97.3 97.3-100 VP3 81.9 97.1 81.5-96.9 97.1-99.2 VP1 82.5 97.2 83.7-97.3 95.7-99.3 2A 80.4 92.0 77.3-94.0 89.3-97.3 2B 79.1 96.0 77.8-81.8 91.9-97.0 2C 80.3 98.8 79.4-84.7 96.4-98.2 3A 69.7 90.9 71.2-78.8 81.8-90.9 3B 83.3 90.9 78.8-87.9 90.9-100 3C 79.2 95.6 76.1-84.3 92.3-97.3 3D 79.1 97.4 79.1-86.1 96.4-98.7 3’UTR 81.9 72.0-95.1 Genome 81.0 81.0-90.6 表 3 135V3/YN/CHN/2022分离株与其他EVB病毒分离株之间的基因组不同区域的一致性比较
Table 3. Consistency comparison of different genomic regions between 135V3/YN/CHN/2022 isolates and other EVB virus isolates
基因区域 毒株 核苷酸一致性(%) 基因登录号 5’UTR CVB2/YN31V3 90.51 MW365443 VP4 CVB2/YN31V3 96.65 MW365443 VP2 CVB2/YN31V3 97.59 MW365443 VP3 CVB2/NODE_5_ 2019-AZ 97.05 OP627804 VP1 CVB2/YN31V3 97.28 MW365443 2A CVB2/YN31V3 94.21 MW365443 2B E6/GHA:CEN:ASE/2017 83.84 MH005791 2C E31/USA/13H4/2016 92.60 OQ791570 3A Env_2017_Jan_CV-B4 89.39 MG451808 3B E9-11-2042-1 92.42 MH144605 3C Env_2017_Jan_CV-B4 89.98 MG451808 3D CVB5/Swab/HB.HS/CHN/2019 89.23 MZ229657 3’UTR E12 / K1529/YN/CHN/2013 95.1 MF083154 genome CVB2/YN31V3 90.62 MW365443 P1 CVB2/YN31V3 97.07 MW365443 P2 E31/USA/13H4/2016 88.72 OQ791570 P3 CVB5/ Swab/HB.HS/CHN/2019 87.64 MZ229657 -
[1] Andino R, Kirkegaard K, MacAdam A, et al. The Picornaviridae family: Knowledge gaps, animal models, countermeasures, and prototype pathogens[J]. J Infect Dis, 2023, 228(Supplement 6): S427-S445. [2] Ushioda W, Kotani O, Kawachi K, et al. Neuropathology in neonatal mice after experimental coxsackievirus B2 infection using a prototype strain, Ohio-1[J]. J Neuropathol Exp Neurol, 2020, 79(2): 209-225. [3] Glenet M, Heng L, Callon D, et al. Structures and functions of viral 5’ non-coding genomic RNA domain-I in group-B enterovirus infections[J]. Viruses, 2020, 12(9): 919. doi: 10.3390/v12090919 [4] Hrebík D, Füzik T, Gondová M, et al. ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating[J]. Proc Natl Acad Sci U S A, 2021, 118(19): e2024251118. doi: 10.1073/pnas.2024251118 [5] Wang S H, Wang K, Zhao K, et al. The structure, function, and mechanisms of action of enterovirus non-structural protein 2C[J]. Front Microbiol, 2020, 11: 615965. [6] Brouwer L, Benschop KSM, Nguyen D, et al. Recombination analysis of non-poliovirus members of the Enterovirus C species; restriction of recombination events to members of the same 3DPol cluster[J]. Viruses, 2020, 12(7): 706. doi: 10.3390/v12070706 [7] Cordeiro e Cunha J, Silva A L, Nogueira R M, et al. Exuberant hand-foot-mouth disease: An immunocompetent adult with atypical findings[J]. Eur J Case Rep Intern Med, 2020, 7(7): 12. doi: 10.12890/2020_001609 [8] 范丽霞, 汪春翔, 赵生仓, 等. 2018年青海省手足口病病原谱的构成及基因特征分析[J]. 中国病原生物学杂志, 2020, 15(11): 1320-1323. doi: 10.13350/j.cjpb.201116 [9] Ahmed R, Moaddab A, Hussain SW, et al. A rare case of dilated cardiomyopathy, focal segmental glomerulosclerosis, and Bell's palsy in a 29-year-old male after coxsackievirus infection[J]. Cureus, 2022, 14(6): e26285. [10] Zhang Q, Yuan J, Zhao W, et al. Coxsackie B virus-induced myocarditis in a patient with a history of lymphoma: A case report and review of literature[J]. Medicine, 2024, 103(10): e37248. doi: 10.1097/MD.0000000000037248 [11] Sousa I P Jr, de Lourdes Aguiar Oliveira M, Burlandy F M, et al. Molecular characterization and epidemiological aspects of non-polio enteroviruses isolated from acute flaccid paralysis in Brazil: A historical series (2005–2017)[J]. Emerg Microbes Infect, 2020, 9(1): 2536-2546. doi: 10.1080/22221751.2020.1850181 [12] Sousa I P Jr, dos Santos F B, de Paula V S, et al. Viral and prion infections associated with central nervous system syndromes in Brazil[J]. Viruses, 2021, 13(7): 1370. doi: 10.3390/v13071370 [13] Jartti M, Flodström-Tullberg M, Hankaniemi M M. Enteroviruses: Epidemic potential, challenges and opportunities with vaccines[J]. J Biomed Sci, 2024, 31: 73. [14] Machado R S, Tavares F N, Sousa I P Jr. Global landscape of coxsackieviruses in human health[J]. Virus Res, 2024, 344: 199367. doi: 10.1016/j.virusres.2024.199367 [15] Cheng C C, Chu P H, Huang H W, et al. Phylodynamic and epistatic analysis of coxsackievirus A24 and its variant[J]. Viruses, 2024, 16(8): 1267. doi: 10.3390/v16081267 [16] 胡亚芳, 朱汝南. 肠道病毒基因重组研究进展[J]. 国际病毒学杂志, 2022, 29(1): 80-84. doi: 10.3760/cma.j.issn.1673-4092.2021.6.017 [17] 刘煜菡, 张名, 许丹菡, 等. RD、KMB-17及Vero细胞对肠道病毒分离培养敏感性的比较[J]. 中国生物制品学杂志, 2024, 37(08): 917-920. doi: 10.13200/j.cnki.cjb.004272 [18] Kalendar R, Shustov AV, Schulman AH, et al. Palindromic sequence-targeted (PST) PCR, version 2: an advanced method for high-throughput targeted gene characterization and transposon display[J]. Front Plant Sci, 2021, 12: 691940. [19] Li F, Wang D, Su F, et al. Epidemiological, etiological, and serological characteristics of hand, foot, and mouth disease in Guizhou Province, Southwest China, from 2008 to 2023[J]. PLoS Negl Trop Dis, 2025, 19(8): e0013394. [20] 杨建辉, 张明瑜, 张璐, 等. 2023年河南省急性弛缓性麻痹病例中非脊髓灰质炎肠道病毒病原学及流行病学分析[J]. 病毒学报, 2025, 41(03): 634-644. doi: 10.13242/j.cnki.bingduxuebao.250027 [21] Zhang Q, Yuan J, Zhao W, et al. Coxsackie B virus-induced myocarditis in a patient with a history of lymphoma: a case report and review of literature[J]. Medicine (Baltimore), 2024, 103(10): e37248. doi: 10.1097/md.0000000000037248 [22] Fratty I S, Kriger O, Weiss L, et al. Molecular analysis of coxsackievirus B2 associated with severe symptoms of the central nervous system[J]. J Med Virol, 2024, 96(11): e70066. doi: 10.1002/jmv.70066 [23] 周冬, 高孟, 高丽美, 等. 重组柯萨奇病毒构建及其感染性研究[J]. 国际流行病学传染病学杂志, 2019, 46(2): 93-98. doi: 10.3760/cma.j.issn.1673-4149.2019.02.001 [24] Ang PY, Chong CWH, Alonso S. Viral Determinants That Drive Enterovirus-A71 Fitness and Virulence[J]. Emerg Microbes Infect, 2021, 10(1): 713-724. doi: 10.1080/22221751.2021.1906754 [25] Yan R, Luo Q, Luo X, et al. Repurposed doxepin targeting host AXL kinase to disrupt viral 2C-mediated immune evasion in Coxsackievirus B infection[J]. Antivir Res, 2025, 244: 106307. doi: 10.1016/j.antiviral.2025.106307 -





 下载:
下载: