Prediction of Shared Target Genes in Cardiac Complications Induced by IAV and SARS-CoV-2 Using Machine Learning and Validation in H1N1 Infection Models
-
摘要:
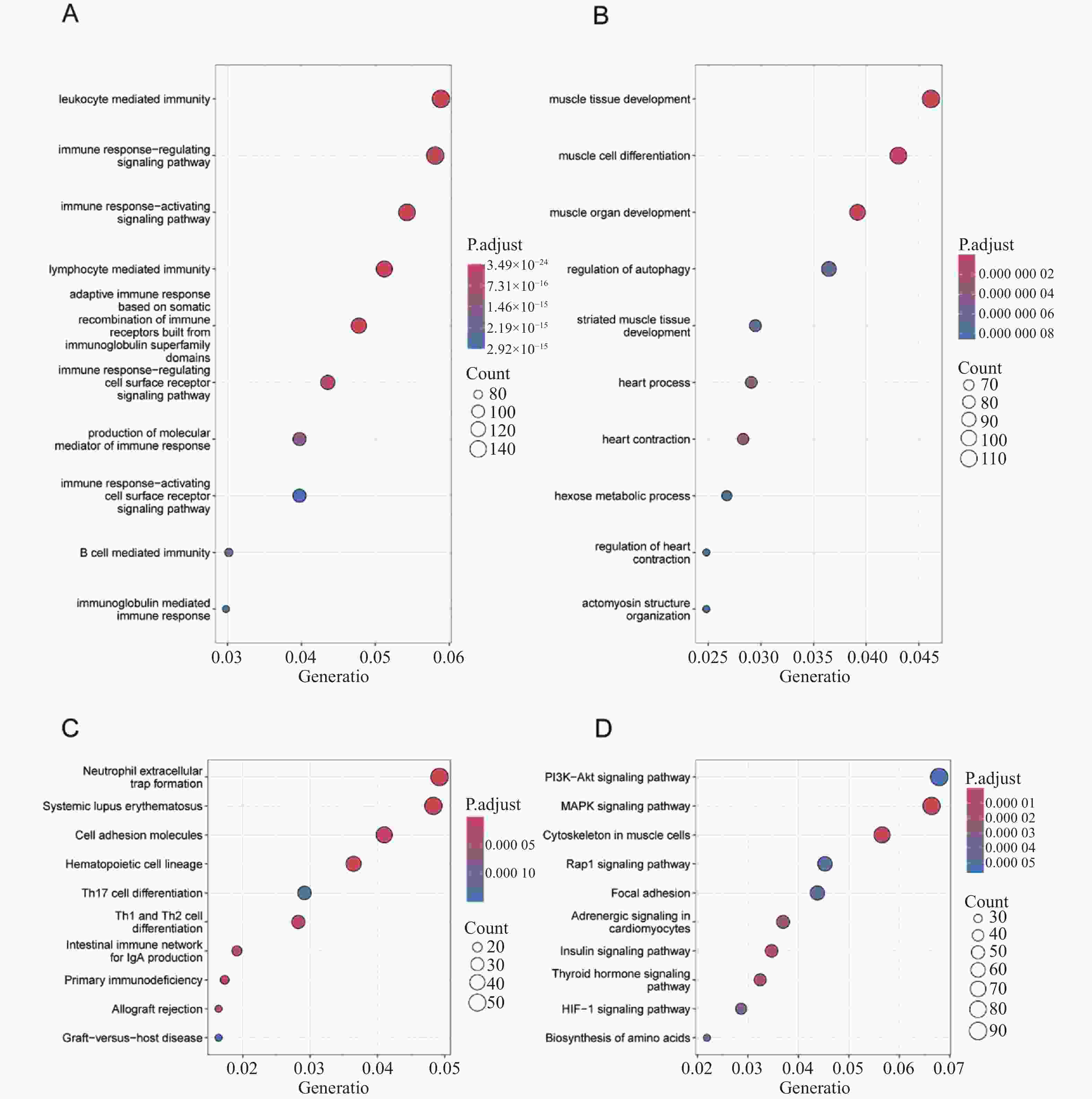
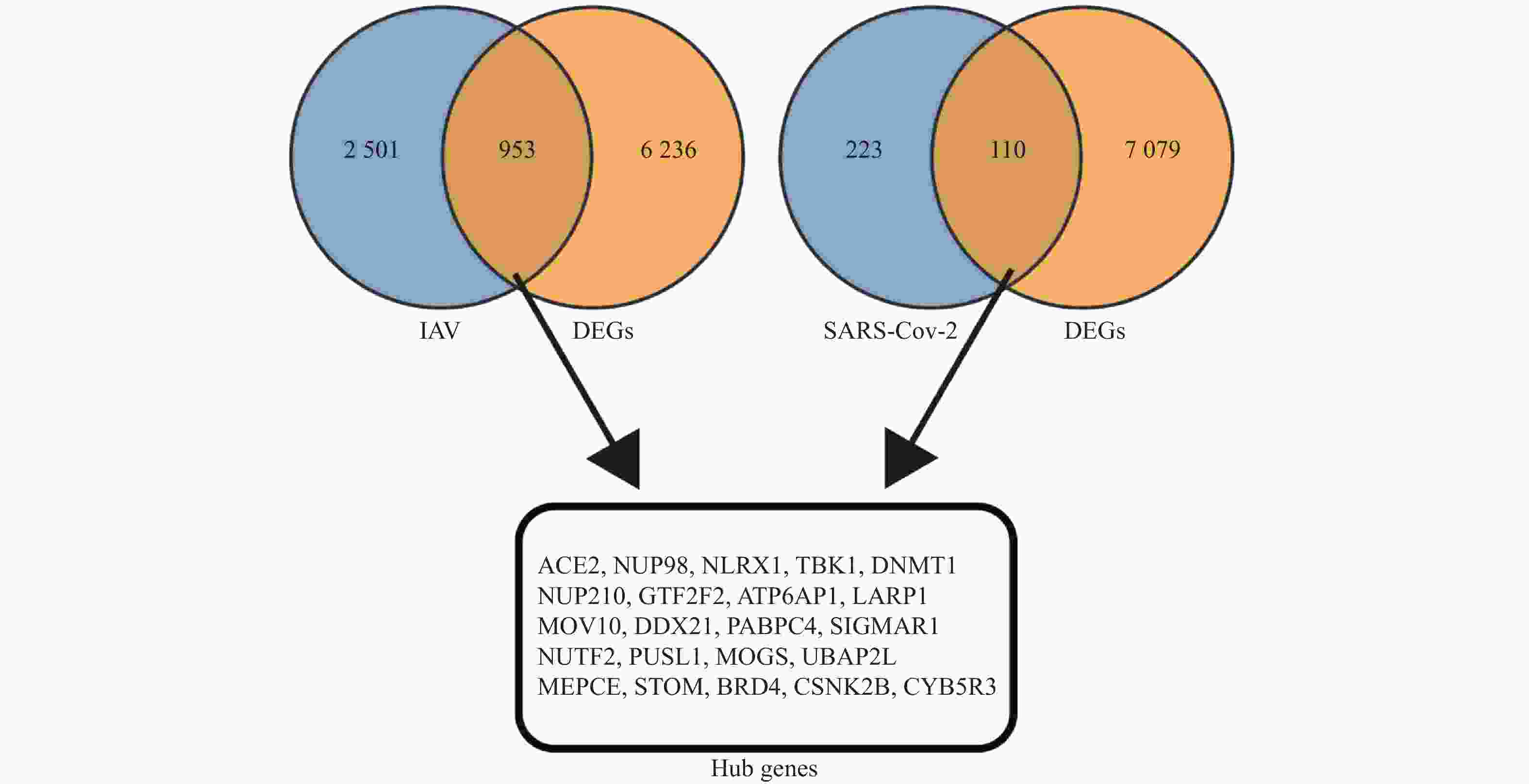
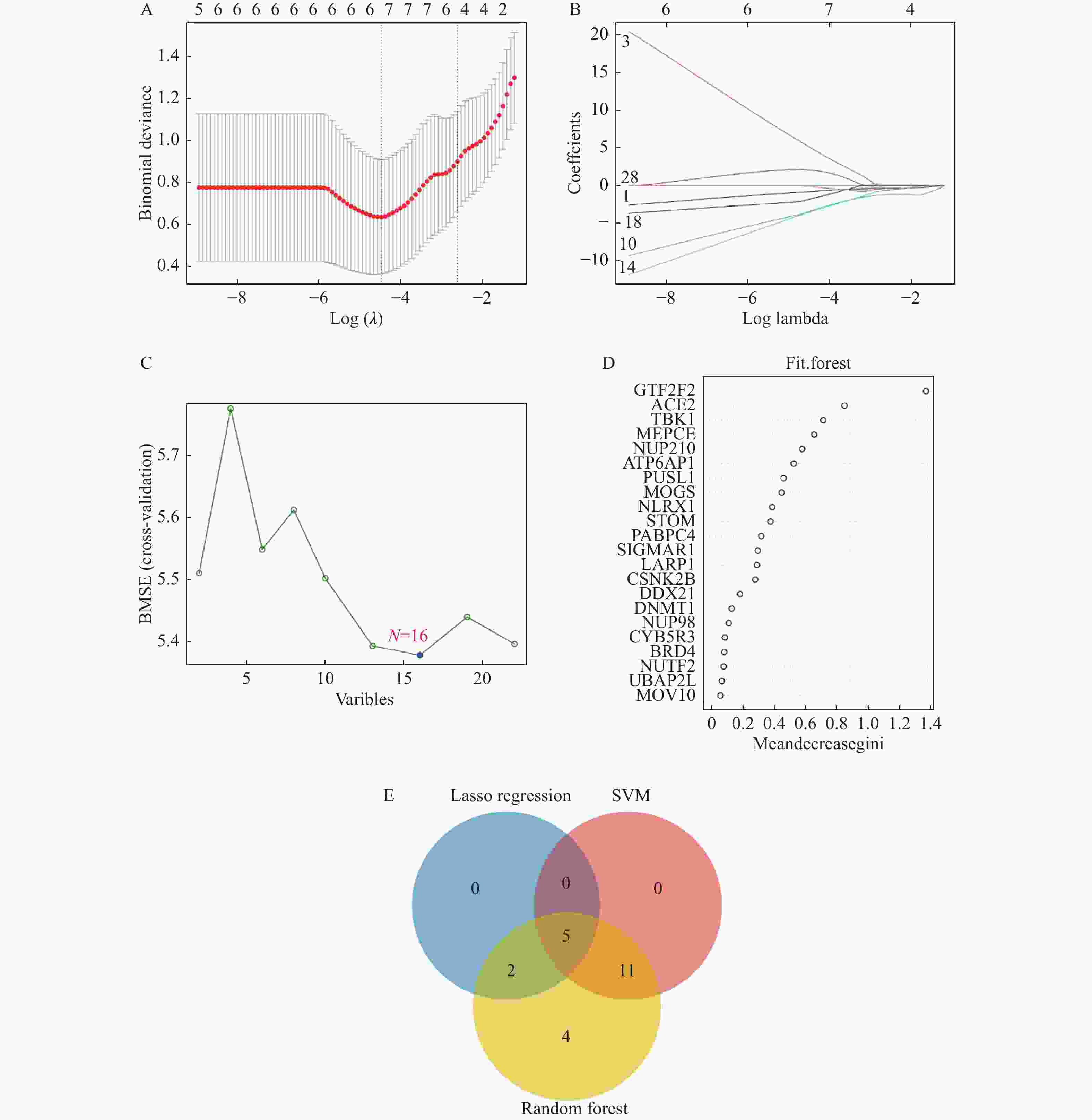
目的 预测并初步验证甲型流感病毒(influenza A virus,IAV)和严重急性呼吸综合征冠状病毒2型(severe acute respiratory syndrome coronavirus 2,SARS-CoV-2)感染引发心脏并发症的共同潜在关键基因。 方法 基于GEO(gene expression omnibus)数据库获取心脏并发症差异表达基因(differentially expressed genes,DEGs),采用分层交集策略,首先分别将心脏并发症相关DEGs与两个独立来源的病毒相关基因集(包括GeneCards中与IAV感染相关的 3454 个人类基因,以及Human Protein Atlas中与SARS-CoV-2相互作用的333个人类蛋白质编码基因)进行交集分析,随后再将这两个交集的结果进行第二轮交集,进一步确定枢纽基因。采用Lasso回归(lasso regression)、随机森林算法(random forest,RF)和支持向量机算法(support vector machine,SVM)3种机器学习算法对枢纽基因进行筛选。本研究主要采用H1N1病毒感染人心肌细胞系(AC16)和IFITM3-/-基因敲除小鼠模型,对预测基因的表达变化进行体外和体内验证。结果 对3个数据库进行生物信息学分析,筛选出22个枢纽基因。使用3种机器学习算法对枢纽基因进行评估,最终筛选出5个共同的关键基因。随后,通过H1N1感染体外培养的人心肌细胞系(AC16),观察到5种基因的转录水平均呈现出动态变化趋势(P < 0.05)。而利用H1N1感染IFITM3-/-基因敲除小鼠的体内实验结果与体外实验结果一致,亦证实这5种基因转录水平均发生动态变化(P < 0.05)。 结论 通过结合生物信息学分析与机器学习算法,本研究筛选出5个与IAV和SARS-CoV-2感染引起心脏并发症相关的共同关键基因,分别为ACE2、TBK1、NUP210、PUSL1和MEPCE。进一步通过H1N1感染模型的体内和体外实验验证,确认了这些基因均与IAV感染引起的心脏并发症相关。 -
关键词:
- 甲型流感病毒 /
- 严重急性呼吸综合征冠状病毒2型 /
- 心脏病 /
- 机器学习
Abstract:Objective To predict and preliminarily validate potential shared key genes involved in cardiac complications caused by influenza A virus (IAV) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections. Methods Differentially expressed genes (DEGs) associated with cardiac complications were obtained from the Gene Expression Omnibus (GEO) database. A hierarchical intersection strategy was applied. First, cardiac complication related DEGs were overlapped with 2 independent virus related gene sets: 3 454 human genes linked to IAV infection in GeneCards and 333 human protein-coding genes interacting with SARS-CoV-2 in the Human Protein Atlas. The 2 overlap results were then intersected to yield 22 hub genes. Lasso regression, random forest (RF) and support vector machine algorithms (SVM) were employed to refine this list. Predicted genes were validated in vitro in H1N1-infected human cardiomyocyte AC16 cells and in vivo in IFITM3 knockout mice challenged with H1N1, assessing transcriptional changes. Results A total of 22 hub genes were identified through integrative bioinformatics analysis. Application of the 3 machine learning algorithms resulted in 5 common key genes: ACE2, TBK1, NUP210, PUSL1, and MEPCE. In vitro infection of AC16 cells with H1N1 revealed dynamic transcriptional changes in all 5 genes post-infection (P < 0.05). In vivo experiments using H1N1-infected IFITM3 knockout mice confirmed the dynamic mRNA expression changes of these 5 genes, consistent with the in vitro results (P < 0.05). Conclusion By combining multilayered bioinformatics analysis with 3 machine learning approaches, 5 common key genes are identified: ACE2, TBK1, NUP210, PUSL1 and MEPCE. Validation in H1N1 infection models confirms their relevance to IAV-induced cardiac complications. -
Key words:
- Influenza A virus /
- SARS-CoV-2 /
- Heart disease /
- Machine learning
-
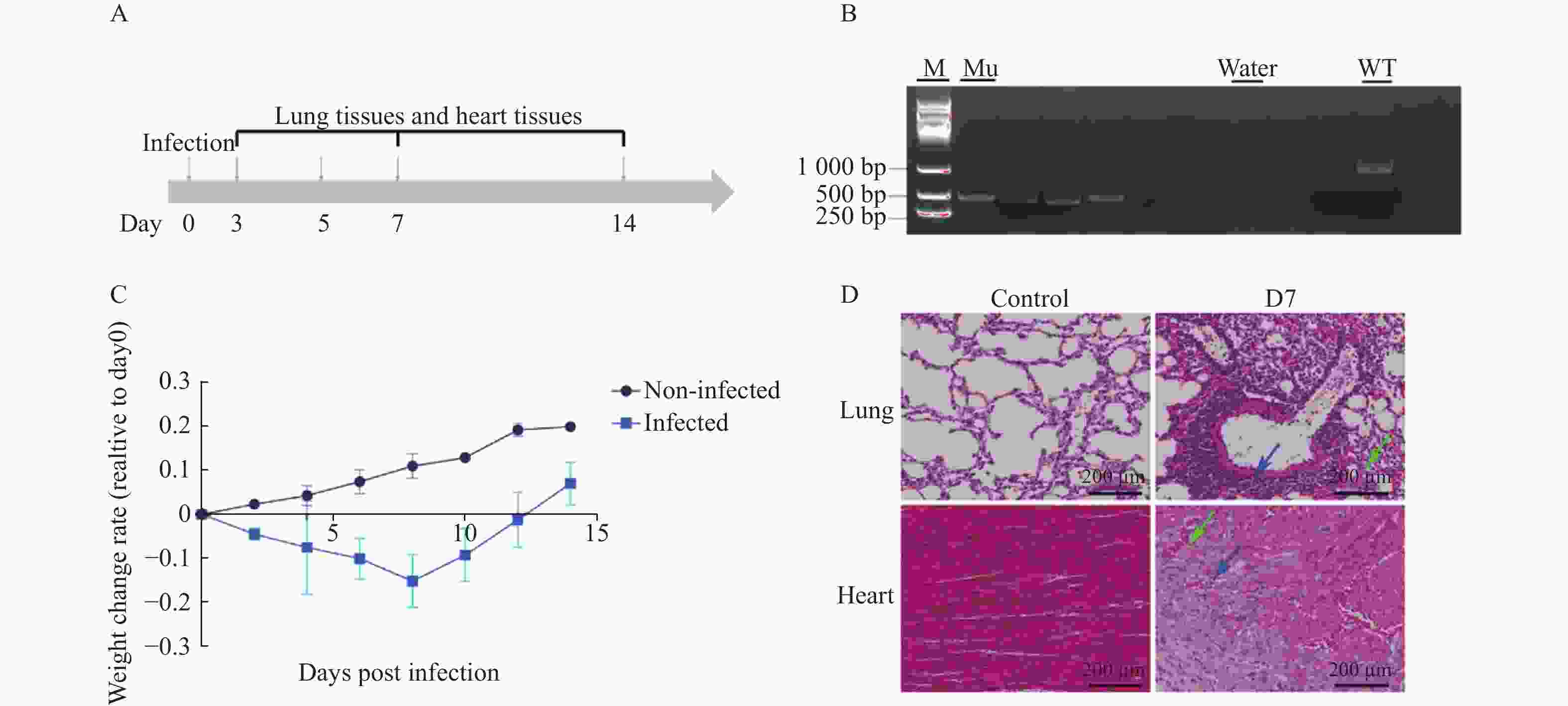
图 6 H1N1感染IFITM3-/-基因敲除小鼠的生理及病理学响应
A:模型小鼠实验设计;B: IFITM3-/-基因敲除小鼠基因型鉴定;C:模型小鼠14天内体重变化率;Non-Infected组为未感染组小鼠,Infected组为感染组小鼠;数据以3次独立重复实验的平均值±标准差(SD)表示;D:实验取材组小鼠感染后肺部及心组织病理变化,蓝色箭头指向炎性细胞浸润区域,绿色箭头指向毛细血管充血区域,标尺刻度为200 µm。
Figure 6. Physiological and pathological responses to H1N1 infection in IFITM3 knockout mice
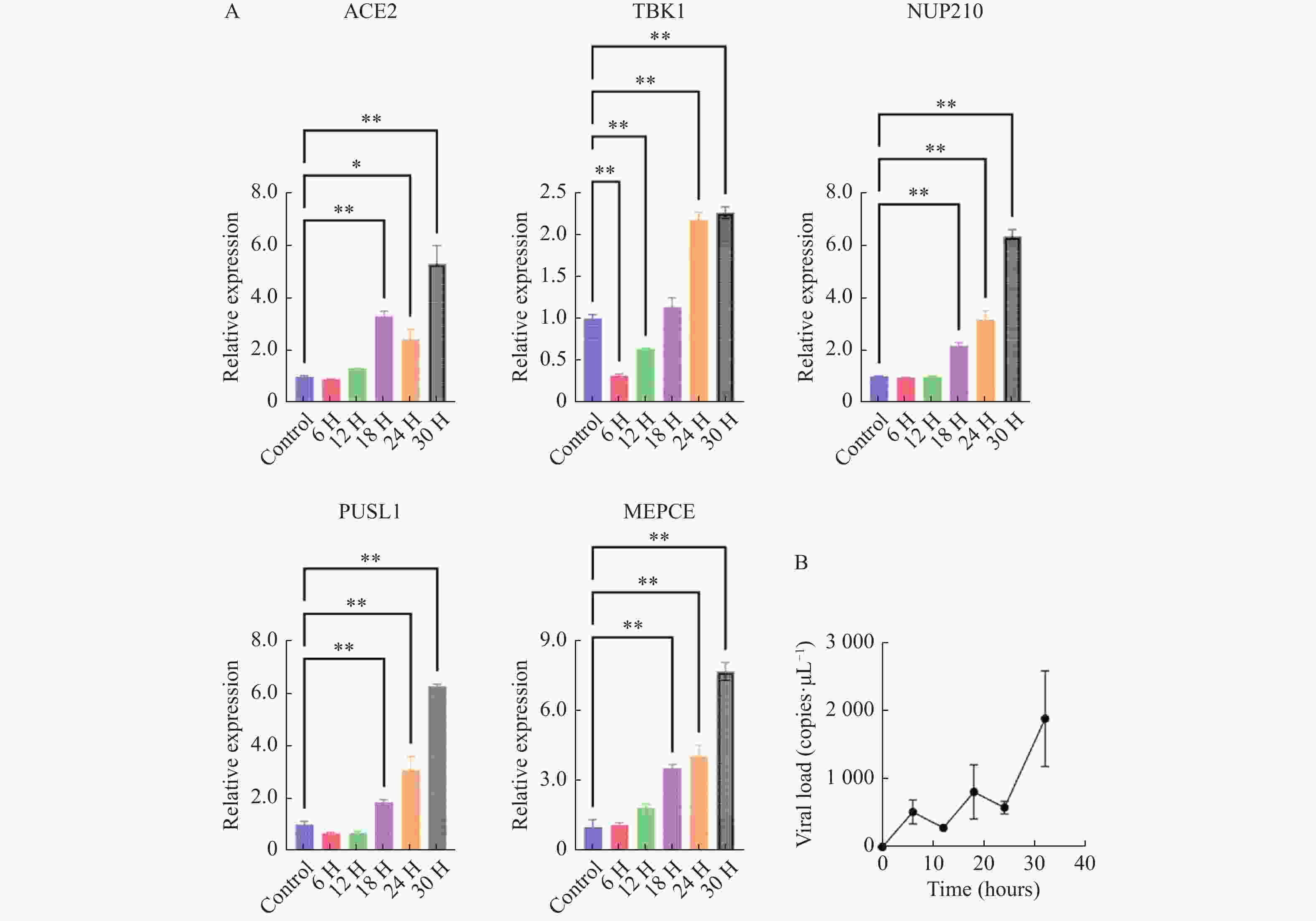
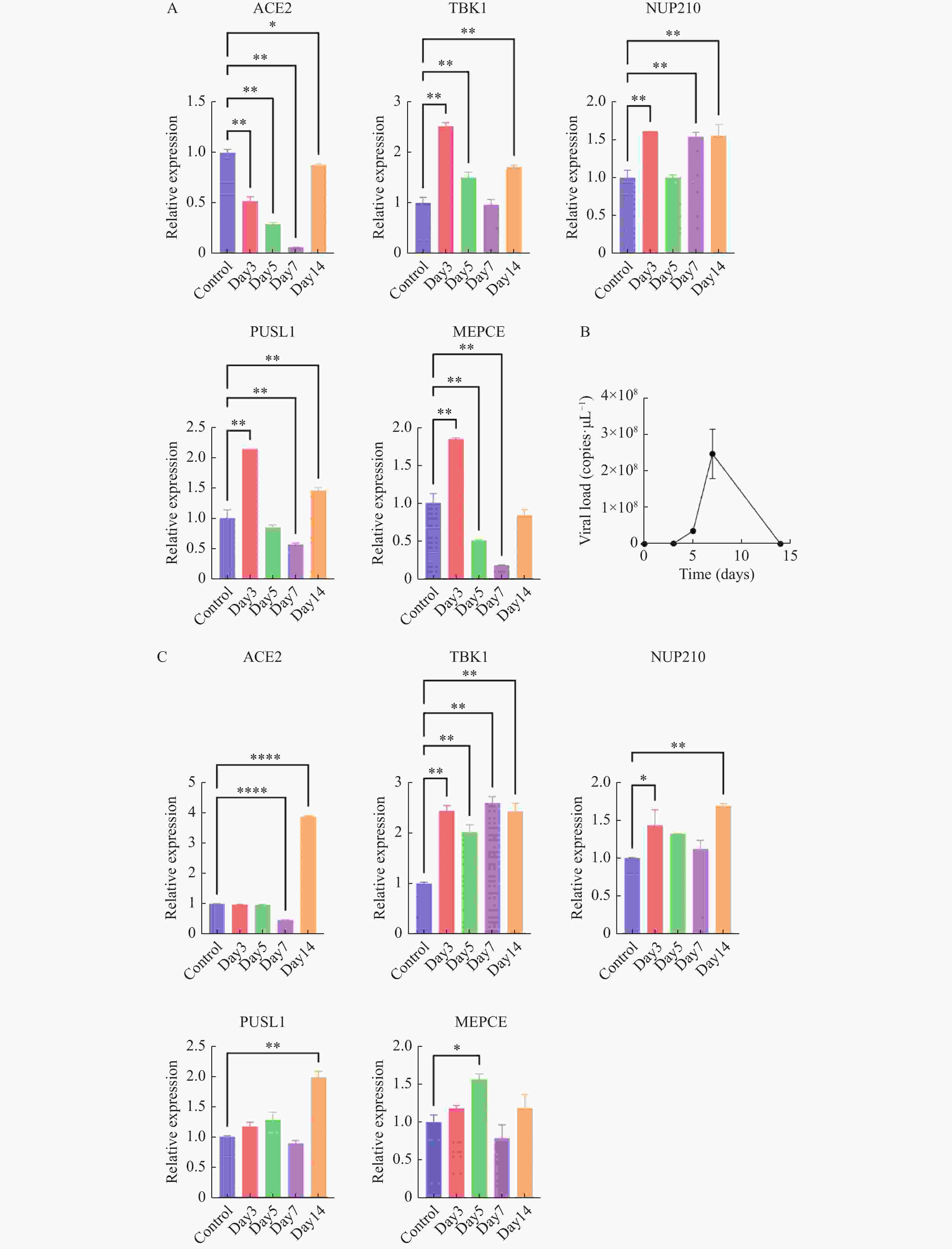
图 7 体内验证H1N1病毒感染IFITM3-/-基因敲除小鼠后特征基因的转录表达水平变化
A:肺组织5个特征基因的转录水平变化;B:肺组织中病毒载量变化;C:心组织中5个特征基因的转录水平变化;数据以3次独立重复实验的平均值±标准差(SD)表示;各组各时间点数据与对照组(未感染流感病毒的IFITM3-/-基因敲除小鼠)进行比较分析,*P < 0.05,**P < 0.01。
Figure 7. In vivo validation of transcriptional expression changes of feature genes following H1N1 virus infection in IFITM3 knockout mice
表 1 PCR引物序列
Table 1. PCR primer sequences
Gene name Sequence (5' to 3') Length
(bp)ACE2(h)-F CGAAGCCGAAGACCTGTTCTA 21 ACE2(h)-R GGGCAAGTGTGGACTGTTCC 20 TBK1(h)-F TGGGTGGAATGAATCATCTACGA 23 TBK1(h)-R GCTGCACCAAAATCTGTGAGT 21 NUP210(h)-F GCACTATCTACGTGGTCGAAC 21 NUP210(h)-R CCTCGAAGAACTCAGCAGGAA 21 PUSL1(h)-F CGTCCCGGACCTACCTGTA 19 PUSL1(h)-R CCTGCATGGCGACCATATCC 20 MEPCE(h)-F CCGCCAAAACATCCGACACTA 21 MEPCE(h)-R CCCGTGACGAAGACAACATTG 21 GAPDH(h)-F TGTGGGCATCAATGGATTTGG 20 GAPDH(h)-R ACACCATGTATTCCGGGTCAAT 21 ACE2(m)-F GGAGCCTGTCAGGGCTACT 19 ACE2(m)-R CCACAAGAATCTGTACCTTCTGC 23 TBK1(m)-F ACTGGTGATCTCTATGCTGTCA 22 TBK1(m)-R TTCTGGAAGTCCATACGCATTG 22 NUP210(m)-F GGGCGCACGATGTTCAGAA 19 NUP210(m)-R CACCACCAGGTCGAAATGGG 20 PUSL1(m)-F GCTCCTGGCCTAATCAACTG 20 PUSL1(m)-R CAGACTGGAAGGCACTAAAATCA 23 MEPCE(m)-F GACCCGCTCAGTCTCAACAC 20 MEPCE(m)-R GGCGTTGCTATCATTCCCTCC 21 GAPDH(m)-F AGGTCGGTGTGAACGGATTTG 21 GAPDH(m)-R TGTAGACCATGTAGTTGAGGTCA 23 FLUAM(m)-1F AAGACCAATCCTGTCACCTCTGA 23 FLUAM(m)-1R CAAAGCGTCTACGCTGCAGTCC 22 FLUAM(m)-1P TTTGTGTTCACGCTCACCGT 20 -
[1] Vos T,Lim S S,Abbafati C,et al. Global burden of 369 diseases and injuries in 204 countries and territories,1990-2019: A systematic analysis for the global burden of disease study 2019[J]. Lancet,2020,396(10258):1204-1222. doi: 10.1016/S0140-6736(20)30925-9 [2] Zhou P,Yang X L,Wang X G,et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin[J]. Nature,2020,579(7798):270-273. doi: 10.1038/s41586-020-2012-7 [3] Jefferson T,Dooley L,Ferroni E,et al. Physical interventions to interrupt or reduce the spread of respiratory viruses[J]. Cochrane Database Syst Rev,2023,1(1):CD006207. [4] Sun H,Xiao Y,Liu J,et al. Prevalent eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection[J]. Proc Natl Acad Sci U S A,2020,117(29):17204-17210. doi: 10.1073/pnas.1921186117 [5] Protonotarios A,Marelli-Berg F. Influenza-associated cardiac injury: a disease of the cardiac conduction system?[J]. Cardiovasc Res,2021,117(3):643-644. doi: 10.1093/cvr/cvaa174 [6] Ramadan M S,Bertolino L,Marrazzo T,et al. Cardiac complications during the active phase of COVID-19: review of the current evidence[J]. Intern Emerg Med,2021,16(8):2051-2061. doi: 10.1007/s11739-021-02763-3 [7] Ouranos K,Vassilopoulos S,Vassilopoulos A,et al. Cumulative incidence and mortality rate of cardiovascular complications due to laboratory-confirmed influenza virus infection: a systematic review and meta-analysis[J]. Rev Med Virol,2024,34(1):e2497. doi: 10.1002/rmv.2497 [8] Dhakal B P,Sweitzer N K,Indik J H,et al. SARS-CoV-2 infection and cardiovascular disease: COVID-19 heart[J]. Heart Lung Circ,2020,29(7):973-987. doi: 10.1016/j.hlc.2020.05.101 [9] Yang W,Liu X,Wang X. The immune system of chicken and its response to H9N2 avian influenza virus[J]. Vet Q,2023,43(1):1-14. [10] López-Valiñas Á,Valle M,Wang M,et al. Vaccination against swine influenza in pigs causes different drift evolutionary patterns upon swine influenza virus experimental infection and reduces the likelihood of genomic reassortments[J]. Front Cell Infect Microbiol,2023,13:1111143. doi: 10.3389/fcimb.2023.1111143 [11] Gouma S,Anderson E M,Hensley S E. Challenges of making effective influenza vaccines[J]. Annu Rev Virol,2020,7(1):495-512. doi: 10.1146/annurev-virology-010320-044746 [12] Skaarup K G,Modin D,Nielsen L,et al. Influenza and cardiovascular disease pathophysiology: strings attached[J]. Eur Heart J Suppl,2023,25(Suppl A):A5-A11. [13] World Health Organization. Number of COVID-19 cases reported to WHO[EB/OL]. (2025-01-05)[2025-01-21]. https://data.who.int/dashboards/co vid19/cases?n=c. [14] Jackson C B,Farzan M,Chen B,et al. Mechanisms of SARS-CoV-2 entry into cells[J]. Nat Rev Mol Cell Biol,2022,23(1):3-20. [15] World Health Organization. Number of COVID-19 cases reported to WHO[EB/OL]. (2025-01-05)[2025-01-21]. https://data.who.int/dashboards/co vid19/cases?n=c. [16] Kopańska M,Barnaś E,Błajda J,et al. Effects of SARS-CoV-2 inflammation on selected organ systems of the human body[J]. Int J Mol Sci,2022,23(8):4178. doi: 10.3390/ijms23084178 [17] Wu L,O'kane A M,Peng H,et al. SARS-CoV-2 and cardiovascular complications: from molecular mechanisms to pharmaceutical management[J]. Biochem Pharmacol,2020,178:114114. doi: 10.1016/j.bcp.2020.114114 [18] Ogah O S,Umuerri E M,Adebiyi A,et al. SARS-CoV 2 infection (Covid-19) and cardiovascular disease in Africa: health care and socio-economic implications[J]. Glob Heart,2021,16(1):18. doi: 10.5334/gh.829 [19] Xie Y,Xu E,Bowe B,et al. Long-term cardiovascular outcomes of COVID-19[J]. Nat Med,2022,28(3):583-590. doi: 10.1038/s41591-022-01689-3 [20] Al-Aly Z,Davis H,Mccorkell L,et al. Long COVID science,research and policy[J]. Nat Med,2024,30(8):2148-2164. doi: 10.1038/s41591-024-03173-6 [21] Liu P P,Blet A,Smyth D,et al. The science underlying COVID-19: implications for the cardiovascular system[J]. Circulation,2020,142(1):68-78. doi: 10.1161/CIRCULATIONAHA.120.047549 [22] Bowe B,Xie Y,Al-Aly Z. Acute and postacute sequelae associated with SARS-CoV-2 reinfection[J]. Nat Med,2022,28(11):2398-2405. doi: 10.1038/s41591-022-02051-3 [23] Wang D,Hu B,Hu C,et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan,China[J]. JAMA,2020,323(11):1061-1069. doi: 10.1001/jama.2020.1585 [24] Wang E Y,Hulme O L,Khurshid S,et al. Initial precipitants and recurrence of atrial fibrillation[J]. Circ Arrhythm Electrophysiol,2020,13(3):e007716. doi: 10.1161/CIRCEP.119.007716 [25] Yue H,Zhang M,Xing L,et al. The epidemiology and clinical characteristics of co-infection of SARS-CoV-2 and influenza viruses in patients during COVID-19 outbreak[J]. J Med Virol,2020,92(11):2870-2873. doi: 10.1002/jmv.26163 [26] Brochet P,Ianni B M,Laugier L,et al. Epigenetic regulation of transcription factor binding motifs promotes Th1 response in Chagas disease cardiomyopathy[J]. Front Immunol,2022,13:958200. doi: 10.3389/fimmu.2022.958200 [27] Kenney A D,Mcmichael T M,Imas A,et al. IFITM3 protects the heart during influenza virus infection[J]. Proc Natl Acad Sci U S A,2019,116(37):18607-18612. doi: 10.1073/pnas.1900784116 [28] Pannucci P,Jefferson S R,Hampshire J,et al. COVID-19-induced myocarditis: pathophysiological roles of ACE2 and toll-like receptors[J]. Int J Mol Sci,2023,24(6):5374. doi: 10.3390/ijms24065374 [29] Chen L,Hao G. The role of angiotensin-converting enzyme 2 in coronaviruses/influenza viruses and cardiovascular disease[J]. Cardiovasc Res,2020,116(12):1932-1936. doi: 10.1093/cvr/cvaa093 [30] Bailey A L,Dmytrenko O,Greenberg L,et al. SARS-CoV-2 infects human engineered heart tissues and models COVID-19 myocarditis[J]. JACC Basic Transl Sci,2021,6(4):331-345. doi: 10.1016/j.jacbts.2021.01.002 [31] Schweitzer K S,Crue T,Nall J M,et al. Influenza virus infection increases ACE2 expression and shedding in human small airway epithelial cells[J]. Eur Respir J,2021,58(1):2003988. doi: 10.1183/13993003.03988-2020 [32] Wu Q,Kumar N,Lafuse W P,et al. Influenza A virus modulates ACE2 expression and SARS-CoV-2 infectivity in human cardiomyocytes[J]. iScience,2022,25(12):105701. doi: 10.1016/j.isci.2022.105701 [33] Bai L,Zhao Y,Dong J,et al. Coinfection with influenza A virus enhances SARS-CoV-2 infectivity[J]. Cell Res,2021,31(4):395-403. doi: 10.1038/s41422-021-00473-1 [34] Sui L,Zhao Y,Wang W,et al. SARS-CoV-2 membrane protein inhibits type I interferon production through ubiquitin-mediated degradation of TBK1[J]. Front Immunol,2021,12:662989. doi: 10.3389/fimmu.2021.662989 [35] Wang X,Jiang L,Wang G,et al. Influenza A virus use of BinCARD1 to facilitate the binding of viral NP to importin α7 is counteracted by TBK1-p62 axis-mediated autophagy[J]. Cell Mol Immunol,2022,19(10):1168-1184. doi: 10.1038/s41423-022-00906-w [36] Sawaged S,Mota T,Piplani H,et al. TBK1 and GABARAP family members suppress Coxsackievirus B infection by limiting viral production and promoting autophagic degradation of viral extracellular vesicles[J]. PLoS Pathog,2022,18(8):e1010350. doi: 10.1371/journal.ppat.1010350 [37] Hagan R S,Torres-Castillo J,Doerschuk C M. Myeloid TBK1 signaling contributes to the immune response to influenza[J]. Am J Respir Cell Mol Biol,2019,60(3):335-345. doi: 10.1165/rcmb.2018-0122OC [38] Sakuma S,Zhu E Y,Raices M,et al. Loss of Nup210 results in muscle repair delays and age-associated alterations in muscle integrity[J]. Life Sci Alliance,2022,5(3):e202101216. doi: 10.26508/lsa.202101216 [39] Ackerman E E,Shoemaker J E. Network controllability-based prioritization of candidates for SARS-CoV-2 drug repositioning[J]. Viruses,2020,12(10):1087. doi: 10.3390/v12101087 [40] Addetia A,Lieberman N a P,Phung Q,et al. SARS-CoV-2 ORF6 disrupts bidirectional nucleocytoplasmic transport through interactions with Rae1 and Nup98[J]. mBio,2021,12(2):e00065-00021. [41] Şenbaş Akyazi B,Pirinçal A,Kawaguchi A,et al. Interaction of influenza A virus NS2/NEP protein with the amino-terminal part of Nup214[J]. Turk J Biol,2020,44(2):82-92. doi: 10.3906/biy-1909-49 [42] Genecards. PUSL1 Gene[EB/OL]. [2025-04-15]. https://www.genecards.org/cgi-bin/carddisp.pl?gene=PUSL1. [43] Shen S,Zhang L S. The regulation of antiviral innate immunity through non-m(6)A RNA modifications[J]. Front Immunol,2023,14:1286820. doi: 10.3389/fimmu.2023.1286820 [44] Morais P,Adachi H,Yu Y T. The critical contribution of pseudouridine to mRNA COVID-19 vaccines[J]. Front Cell Dev Biol,2021,9:789427. doi: 10.3389/fcell.2021.789427 [45] Wang Y,Traugot C M,Bubenik J L,et al. N(6)-methyladenosine in 7SK small nuclear RNA underlies RNA polymerase II transcription regulation[J]. Mol Cell,2023,83(21):3818-3834. doi: 10.1016/j.molcel.2023.09.020 [46] Li Z,Fang X,Zhao B,et al. Liquid-liquid phase separation of LARP7 restrains HIV-1 replication[J]. EMBO Rep,2025,26(8):1935-1956. doi: 10.1038/s44319-025-00421-9 -





 下载:
下载: