Ferroptosis-Related LncRNAs Signature Predicts the Prognosis of Stomach Adenocarcinoma
-
摘要:
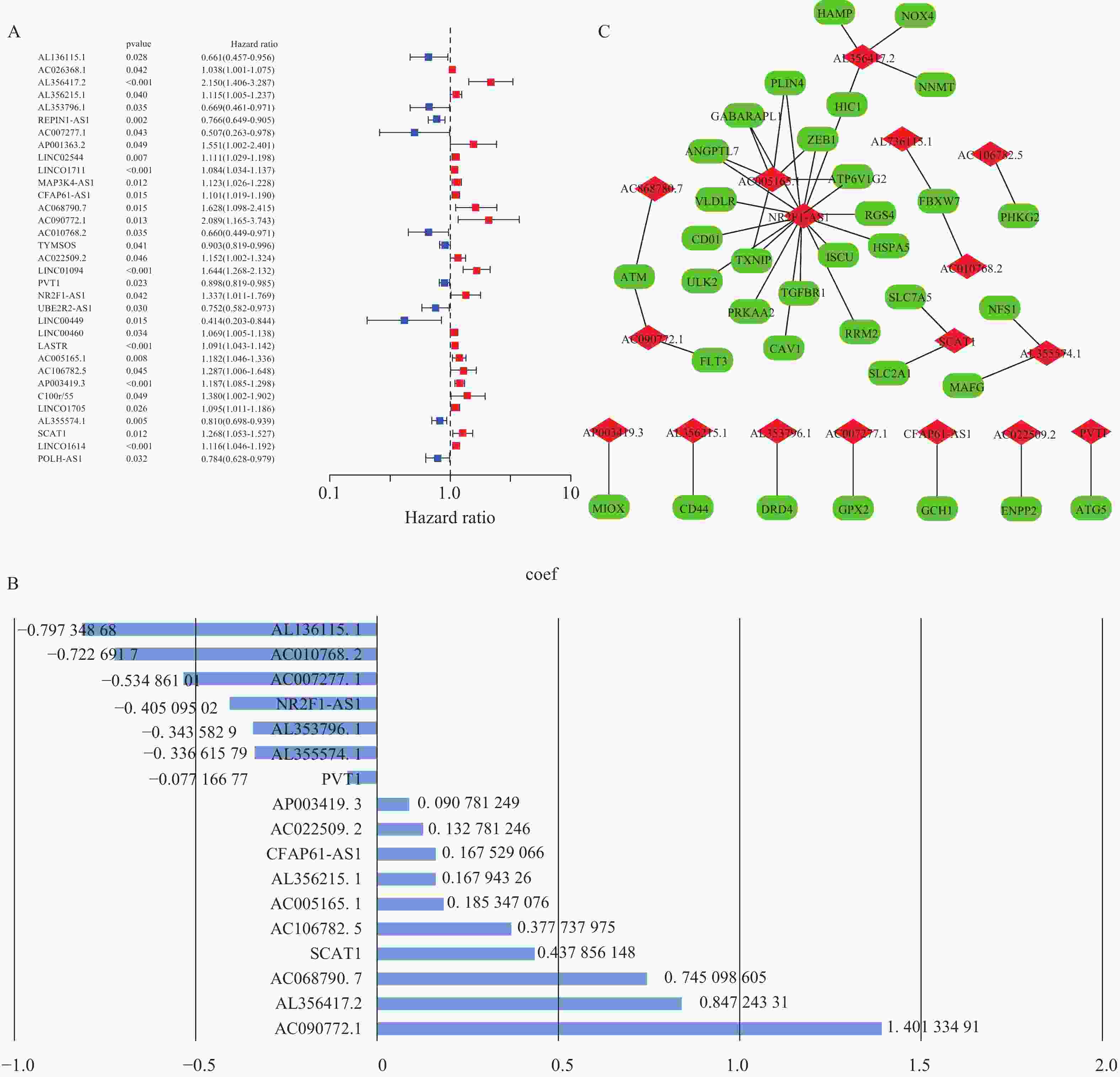
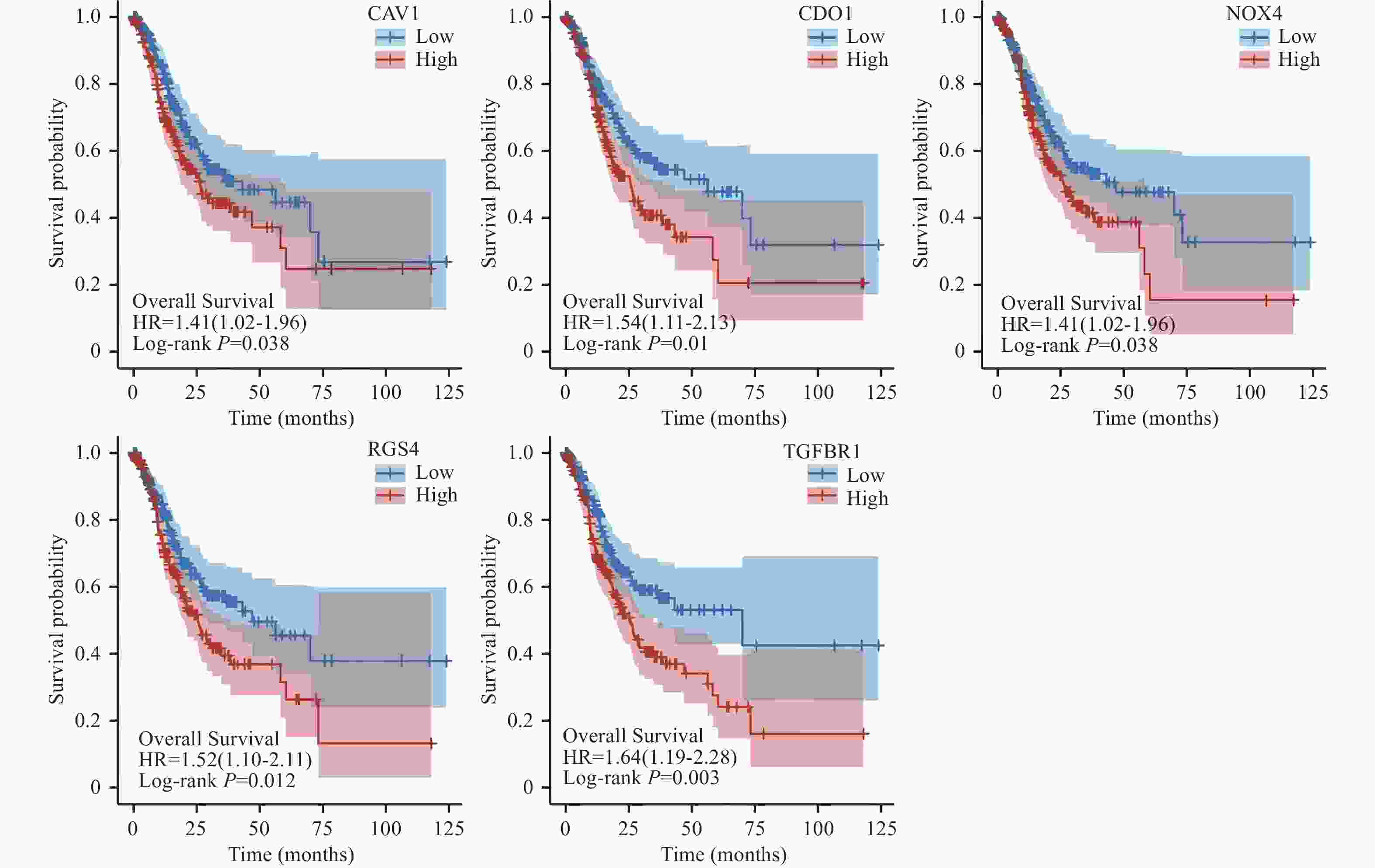
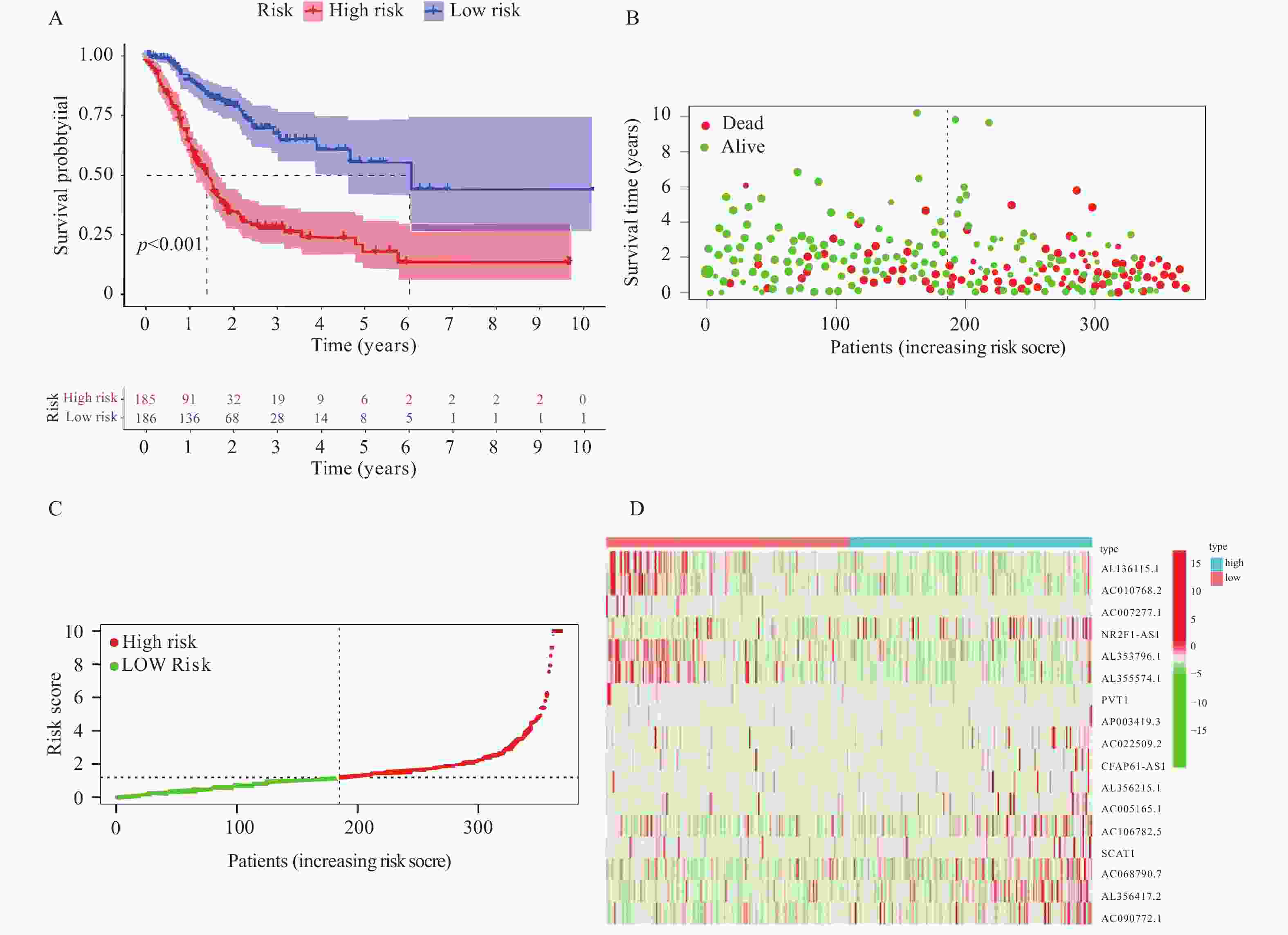
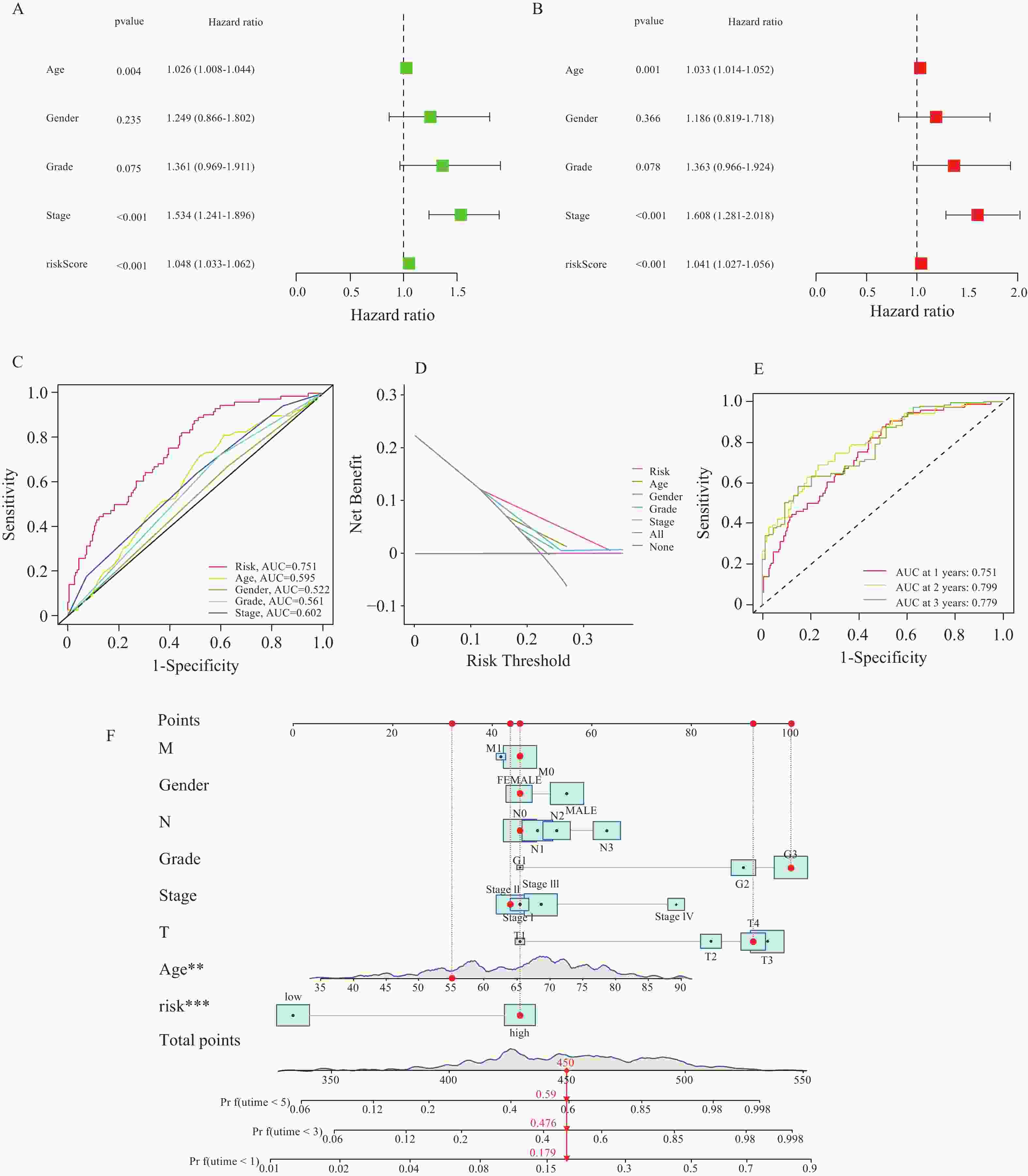
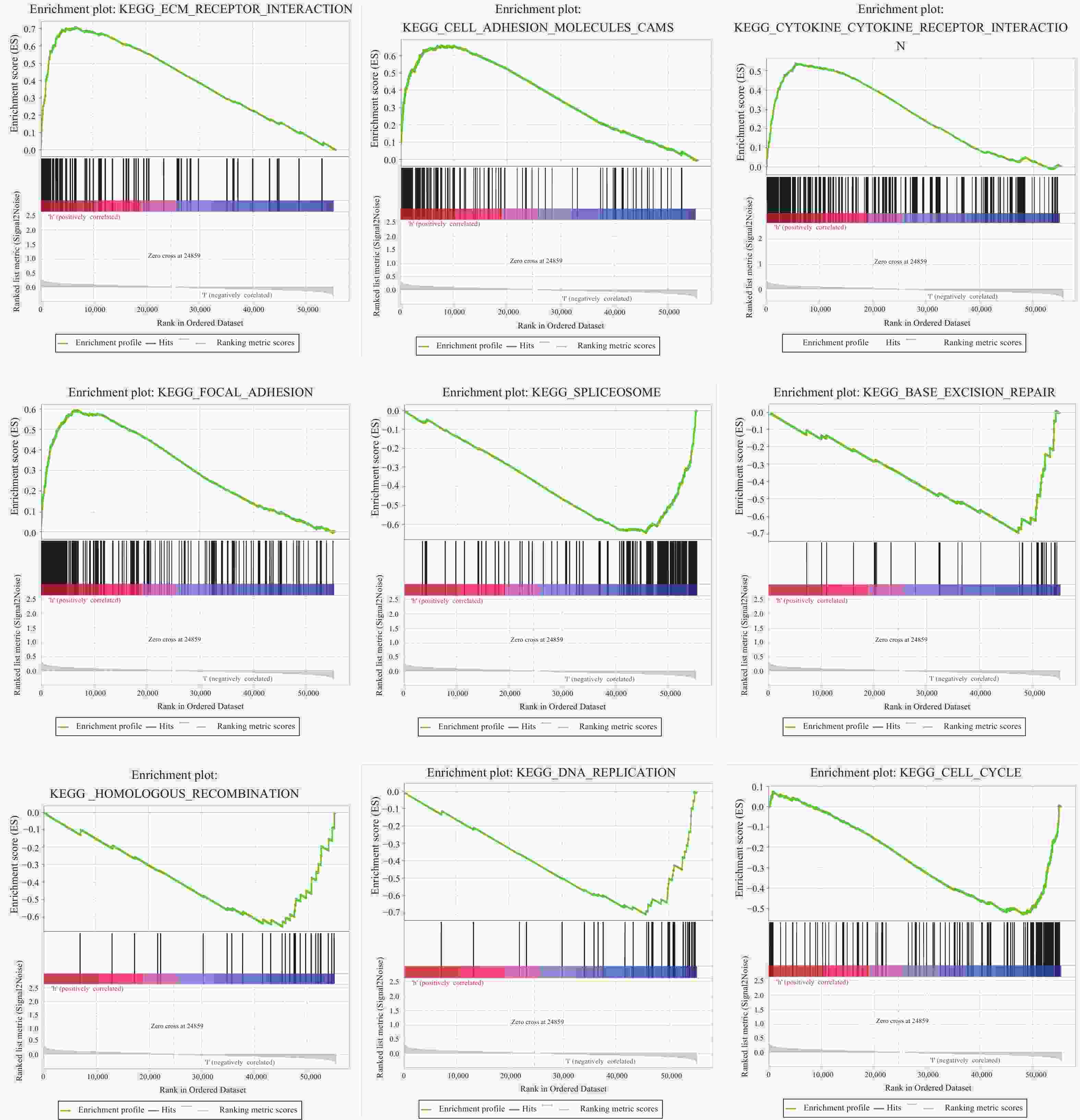
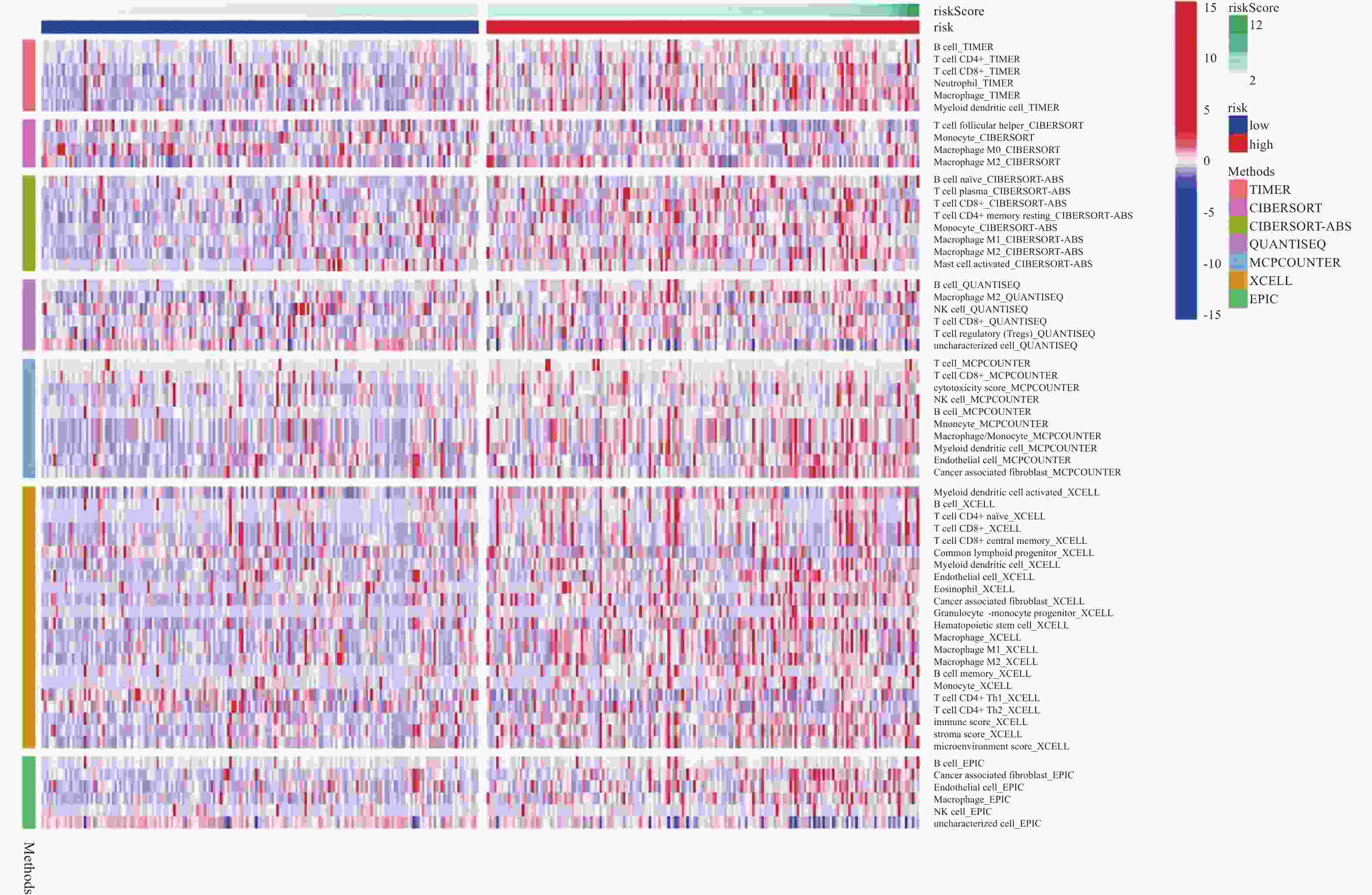
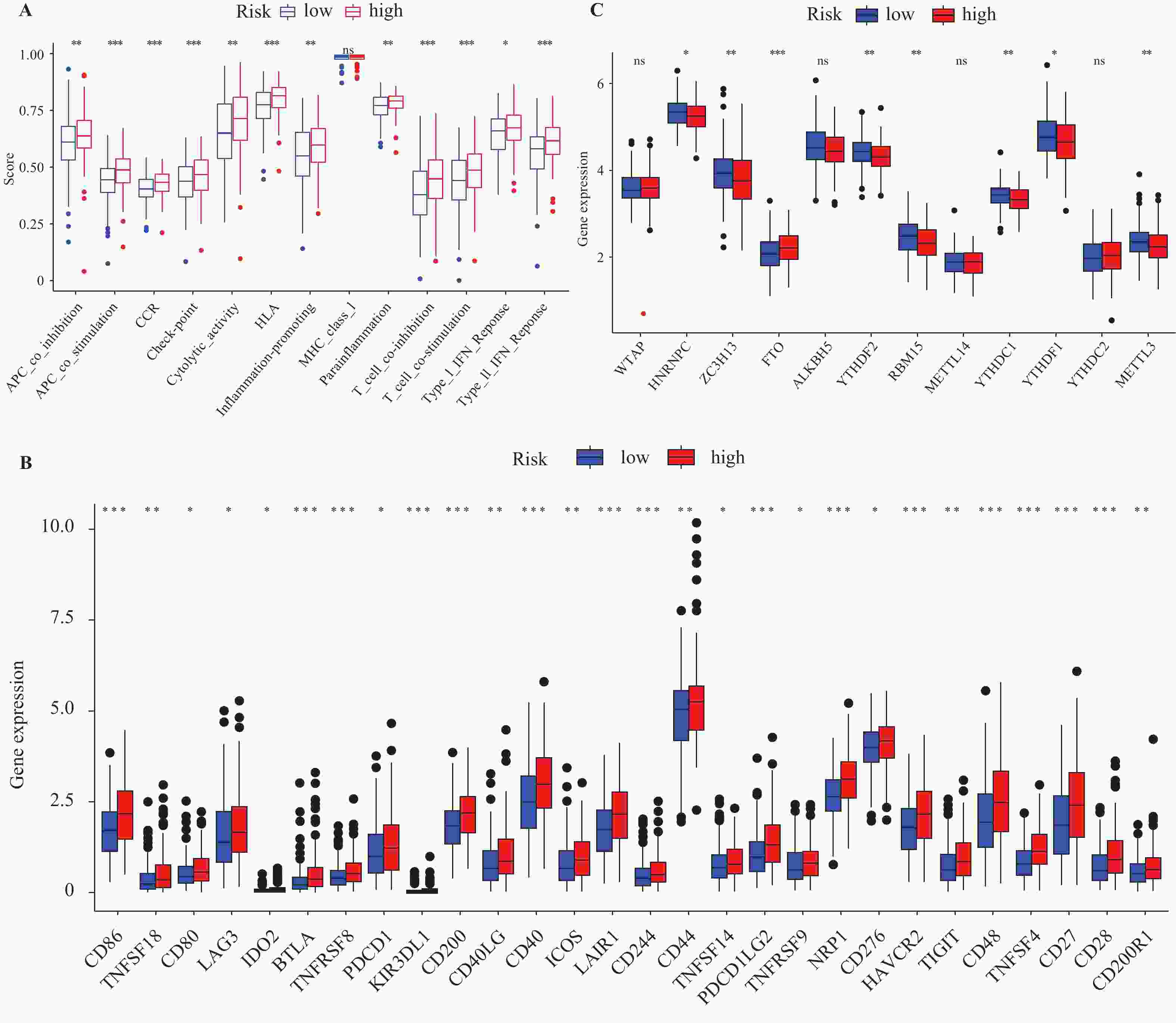
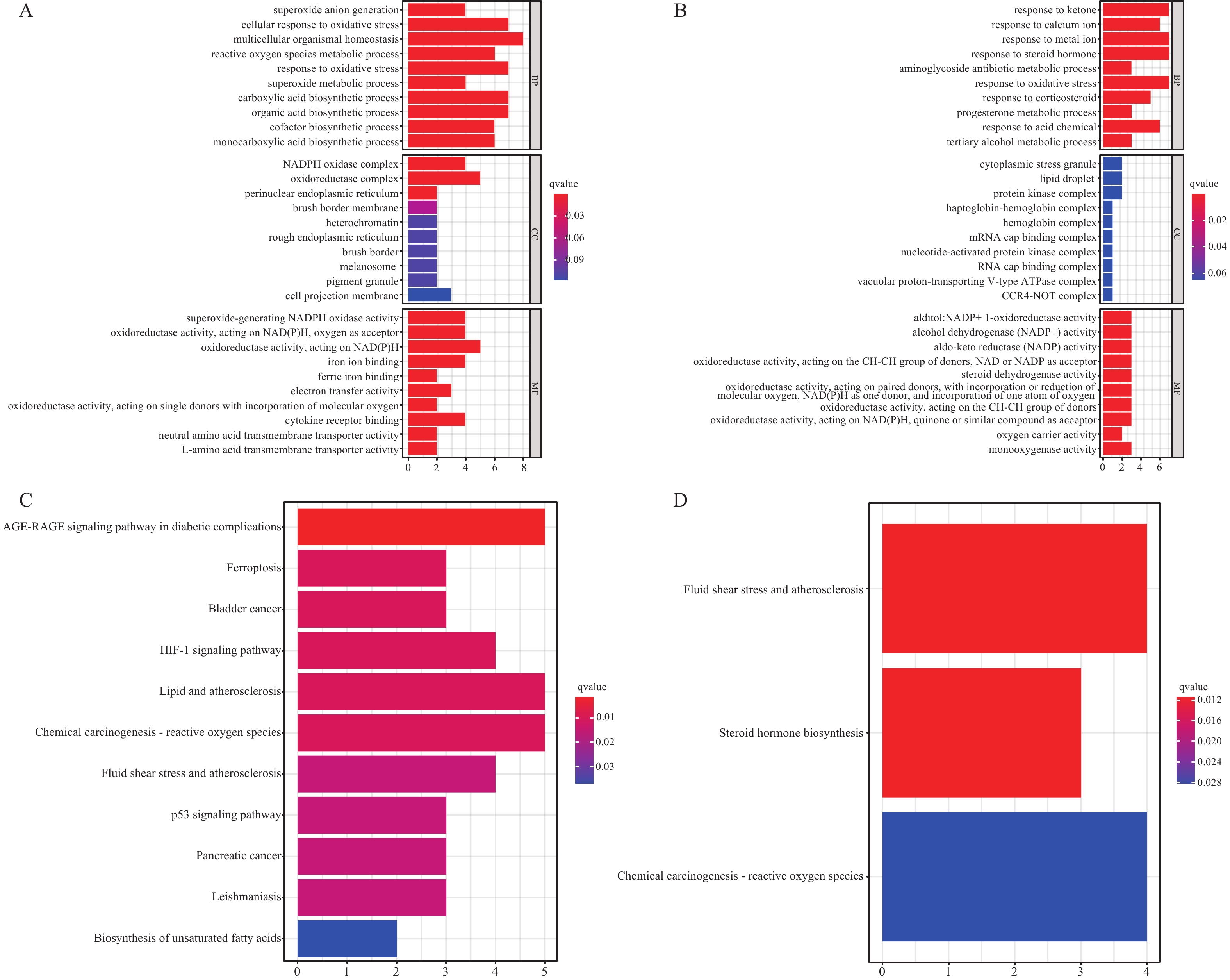
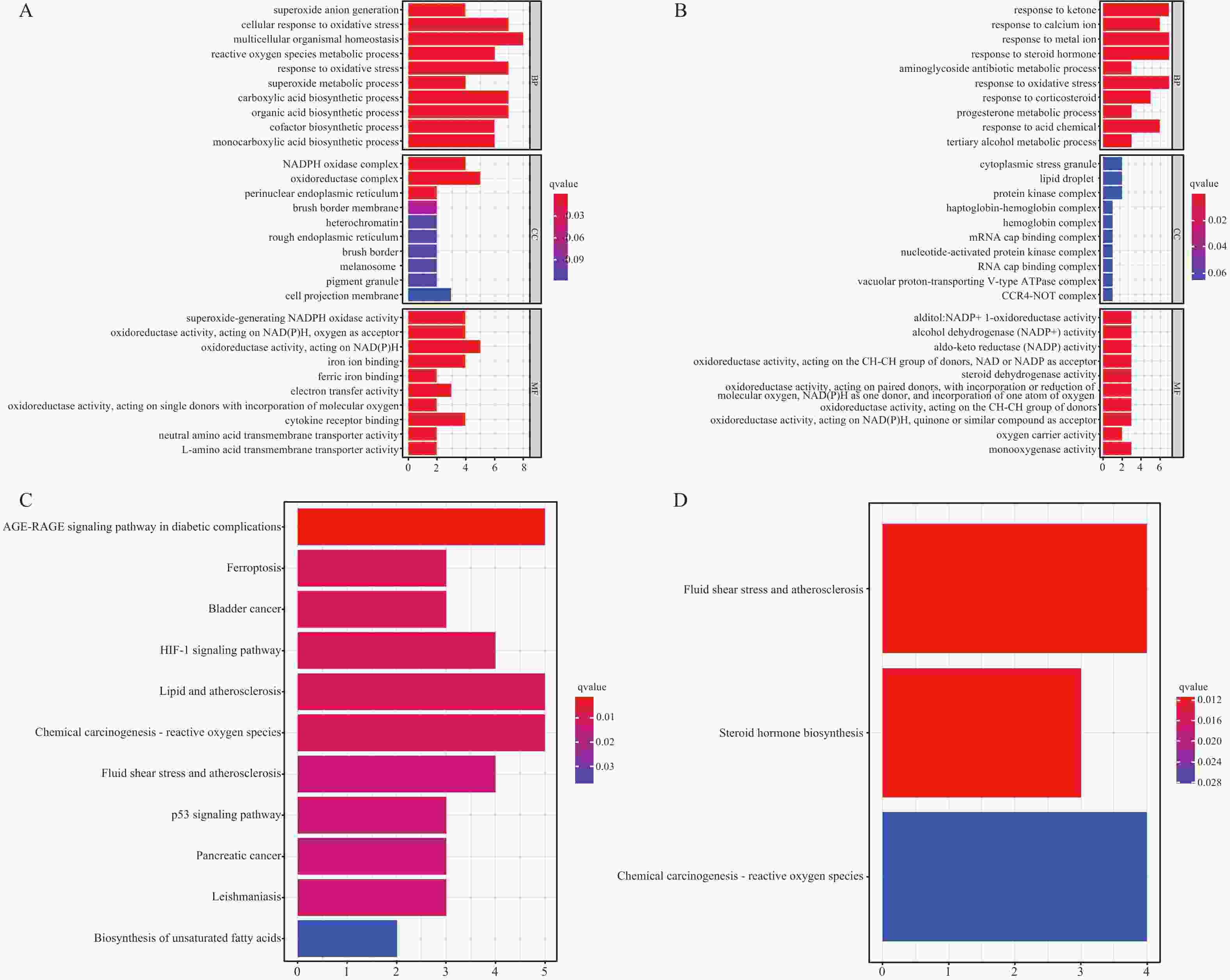
目的 研究胃癌细胞铁死亡相关LncRNA,建立预测胃腺癌患者的生存预后情况的预后模型,为其生物标志物与治疗靶点的开发提供理论依据。 方法 对 TCGA数据库中的胃腺癌患者的转录本测序数据进行分析,并与铁死亡相关基因取交集,通过共表达和差异分析方法,从而筛选出与铁死亡相关的 LncRNA。采用单因素和多因素 Cox回归分析,筛选出与胃腺癌患者预后相关的 LncRNA,从而建立预后评分模型。在此基础上,对每一样本进行风险值计算,并对模型的可靠性进行充分验证;根据模型结果对高低风险组之间进行免疫浸润与免疫反应等差异分析。 结果 肿瘤组织中相较正常组织筛选到与铁死亡相关的503个 LncRNA (上调431个,下调72个);单因素Cox回归分析得出33个可作为独立风险因子的 LncRNA,而多因素Cox回归分析构建出一个由17个 LncRNA组成的预测模型。生存曲线表明高风险的患者比低高风险的患者的存活率显著降低(P < 0.001);单因素和多因素独立预后分析表明,年龄、分期与风险值是患者的独立危险因素;时间依赖的ROC曲线提示,模型的1,2,3年生存率预测AUC值为0.751,0.799,0.779,证明模型具备可靠与稳定性。高低风险组间多个免疫激活反应、免疫细胞的浸润程度与免疫检查点的表达水平存在显著差异(P < 0.001)。 结论 以铁死亡相关lncRNA为基础,建立胃腺癌患者预后预测模型,可较好地评估患者预后情况,纳入模型的LncRNA具备开发为生物标志物与治疗靶点的可行性。 Abstract:Objective To establish a prognostic model that predicts the survival and prognosis of gastric adenocarcinoma patients by studying the LncRNAs related to iron death in gastric cancer cells, thereby providing a theoretical basis for the development of their biomarkers and therapeutic targets. Methods The transcript sequencing data of gastric adenocarcinoma patients in the TCGA database were analyzed and intersected with iron death-related genes, which were screened for iron death-related LncRNAs by co-expression and differential analysis methods. One-way and multifactorial Cox regression analyses were used to screen out the prognostic-related LncRNAs in gastric adenocarcinoma patients, so as to establish the prognostic scoring models. On this basis, risk values were calculated for each sample, and the reliability of the model was fully verified. According to the model results, differences in the immune infiltration and immune response between the high- and low-risk groups were analyzed. Results Tumor tissues were screened for 503 LncRNAs (431 up-regulated and 72 down-regulated) associated with iron death compared to the normal tissues; univariate Cox regression analysis yielded 33 LncRNAs that could be used as the independent risk factors, whereas multivariate Cox regression analysis constructed a predictive model consisting of 17 LncRNAs. Survival curves indicated that patients with the high risk had the significantly lower survival rates than those with the low risk (P < 0.001). Unifactorial and multifactorial independent prognostic analyses showed that age, stage, and risk value were independent risk factors for patients; Time-dependent ROC curves suggested that the predicted AUC values of the model's 1-, 2-, and 3-year survival rates were 0.751, 0.799, and 0.779 respectively, proving that the model was reliable and stable. There were significant differences in multiple immune activation responses, the degree of immune cell infiltration, and the expression levels of immune check points between the high- and low-risk groups. Conclusion The established prognostic prediction model based on iron death-related lncRNAs for gastric adenocarcinoma patients can better assess the prognosis of patients, and the lncRNAs included in the model have the feasibility of being developed into biomarkers and therapeutic targets. -
Key words:
- Stomach adenocarcinoma /
- Ferroptosis /
- LncRNA /
- Prognostic model
-
表 1 患者临床基线资料表[n(%)]
Table 1. The clinical characteristics of patients in the TCGA database [n(%)]
临床特征 分类 人数 死亡人数 n 375 150 性别 女性 134 (35.7) 51(34.0) 男性 241 (64.3) 99(66.0) 年龄(岁) ≤65 173 (46.1) 60(40.0) >65 202 (53.9) 90(60.0) 组织学分级 G1 10 (2.7) 3(2.0) G2 131 (34.9) 49(32.7) G3 234 (62.4) 98(65.3) 病理分期 Stage I 46 (12.3) 11(7.3) Stage II 123 (32.8) 34(22.7) Stage III 165 (44.0) 79(52.7) Stage IV 41 (10.9) 26(17.3) T 分期 T1 15 (4.0) 1(0.7) T2 80 (21.3) 27(18.0) T3 179 (47.8) 79(52.7) T4 101 (26.9) 43(28.6) N 分期 N0 114 (30.4) 29(19.3) N1 102 (27.2) 43(28.7) N2 75 (20.0) 31(20.7) N3 84 (22.4) 47(31.3) M 分期 M0 339 (90.4) 133(88.7) M1 36 (9.6) 17(11.3) -
[1] Bray F,Laversanne M,Weiderpass E,et al. The ever-increasing importance of cancer as a leading cause of premature death worldwide[J]. Cancer,2021,127(16):3029-3030. doi: 10.1002/cncr.33587 [2] Sung H,Ferlay J,Siegel R L,et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA: A Cancer Journal for Clinicians,2021,71(3):209-249. doi: 10.3322/caac.21660 [3] Dixon S J,Lemberg K M,Lamprecht M R,et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death[J]. Cell,2012,149(5):1060-1072. doi: 10.1016/j.cell.2012.03.042 [4] Bystrom L M,Rivella S. Cancer cells with irons in the fire[J]. Free Radical Biology and Medicine,2015,79(5):337-342. [5] Fu D,Wang C,Yu L,et al. Induction of ferroptosis by ATF3 elevation alleviates cisplatin resistance in gastric cancer by restraining Nrf2/Keap1/xCT signaling[J]. Cellular & Molecular Biology Letters,2021,26(1):26. [6] Ma R,Shimura T,Yin C,et al. Antitumor effects of Andrographis via ferroptosis-associated genes in gastric cancer[J]. Oncology Letters,2021,22(1):523. doi: 10.3892/ol.2021.12784 [7] Fatica A,Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development[J]. Nature Reviews Genetics,2014,15(1):7-21. doi: 10.1038/nrg3606 [8] Zhang K,Shi H,Xi H,et al. Genome-wide lncRNA microarray profiling identifies novel circulating lncRNAs for detection of gastric cancer[J]. Theranostics,2017,7(1):213-227. doi: 10.7150/thno.16044 [9] Fu J,Zhao W,Guo D,et al. LncRNA E2F-mediated cell proliferation enhancing LncRNA regulates cancer cell behaviors and affects prognosis of gastric cancer[J]. Digestive Diseases and Sciences,2020,65(5):1348-1354. doi: 10.1007/s10620-019-05855-5 [10] Su X,Zhang J,Luo X,et al. LncRNA LINC01116 promotes cancer cell proliferation,migration and invasion in gastric cancer by positively interacting with LncRNA CASC11[J]. OncoTargets and Therapy,2019,12(3):8117-8123. doi: 10.2147/OTT.S208133 [11] Hong L,Wang H,Wang J,et al. LncRNA PTCSC3 inhibits tumor growth and cancer cell stemness in gastric cancer by interacting with LncRNA linc-pint[J]. Cancer Management and Research,2019,11(11):10393-10399. doi: 10.2147/CMAR.S231369 [12] Zhang Y,Guo S,Wang S,et al. LncRNA OIP5-AS1 inhibits ferroptosis in prostate cancer with long-term cadmium exposure through miR-128-3p/SLC7A11 signaling[J]. Ecotoxicology and Environmental Safety,2021,220(9):112376. doi: 10.1016/j.ecoenv.2021.112376 [13] Lu J,Xu F,Lu H. LncRNA PVT1 regulates ferroptosis through miR-214-mediated TFR1 and p53[J]. Life Sciences,2020,260(1):118305. doi: 10.1016/j.lfs.2020.118305 [14] Yang C,Liu Z,Chang X,et al. NR2F1‐AS1 regulated miR‐423‐5p/SOX12 to promote proliferation and invasion of papillary thyroid carcinoma[J]. Journal of Cellular Biochemistry,2020,121(2):2009-2018. doi: 10.1002/jcb.29435 [15] Bai M,Wu Z,Hyang Y,et al. STAT3 activates the transcription of LncRNA NR2F1-AS1 to promote progression of melanoma via regulating miR3/GOLM1 axis[J]. The Journal of Gene Medicine,2021,23(8):e3338. [16] Wang J,Dong S,Zhang J,et al. LncRNA NR2F1-AS1 regulates miR-371a-3p/TOB1 axis to suppress proliferation of colorectal cancer cells[J]. Cancer Biotherapy & Radiopharmaceuticals,2020,35(10):760-764. [17] Cui Y,Liu L,Wang L,et al. LncRNA PVT1 regulates VEGFC through inhibiting miR‐128 in bladder cancer cells [J]. Journal of Cellular Physiology,2019,234(2): 1346-1353. [18] Yang Y,Zang S,Zhong C,et al. Increased expression of the LncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer[J]. International Journal of Clinical and Experimental Pathology,2014,7(10):6929-6935. [19] Ali M M,Akhade V,Skosalai S T,et al. PAN-cancer analysis of S-phase enriched LncRNAs identifies oncogenic drivers and biomarkers[J]. Nature Communications,2018,9(1):883. doi: 10.1038/s41467-018-03265-1 [20] Guo Y,Zhu T,Guo W,et al. Aberrant CpG island shore region methylation of CAV1 is associated with tumor progression and poor prognosis in gastric cardia adenocarcinoma[J]. Arch Med Res,2016,47(6):460-470. [21] Vwdeld H,Andresen K,Eilertesn I,et al. The novel colorectal cancer biomarkers CDO1,ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers[J]. Int J Cancer,2015,136(4):844-853. [22] Yuan H,LI P P,Zhang L M. The expression of NOX4 gene in stomach adenocarcinoma tissues and its prognostic significance utilizing TCGA database[J]. Xian Dai Zhong Liu Yi Xue,2019,27(17):2995-2999. [23] Guda M,Velpula K,Asuthkar S,et al. Targeting RGS4 ablates glioblastoma proliferation[J]. Int J Mol Sci,2020,721(9):3300. [24] Wang J,Xiang H,Lu Y,et al. Role and clinical significance of TGF‑β1 and TGF‑βR1 in malignant tumors (Review)[J]. Int J Mol Med,2021,47(4):55. -





 下载:
下载: