Research Progress of Indole in E. coli Biofilms on the Surface of Biomaterials
-
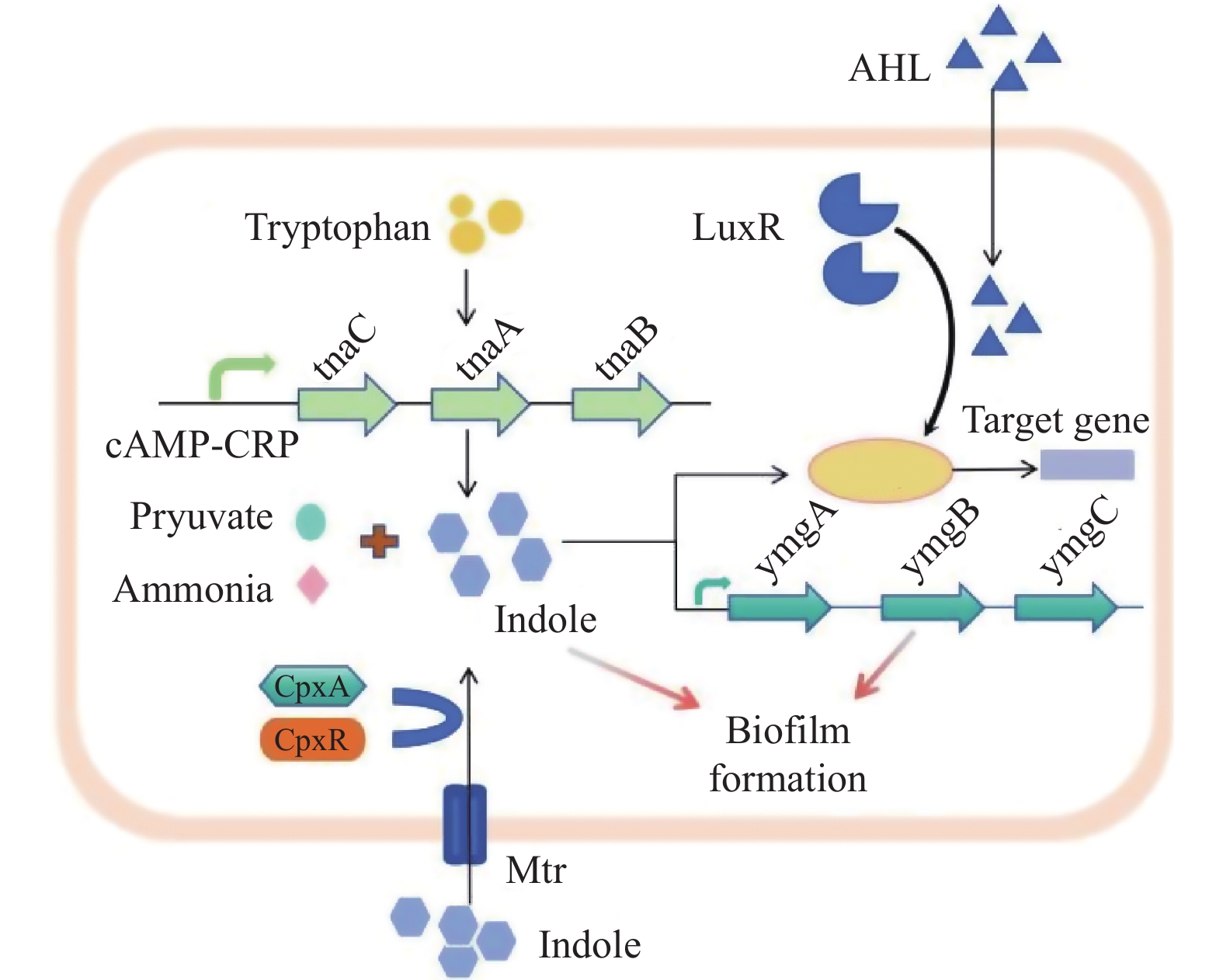
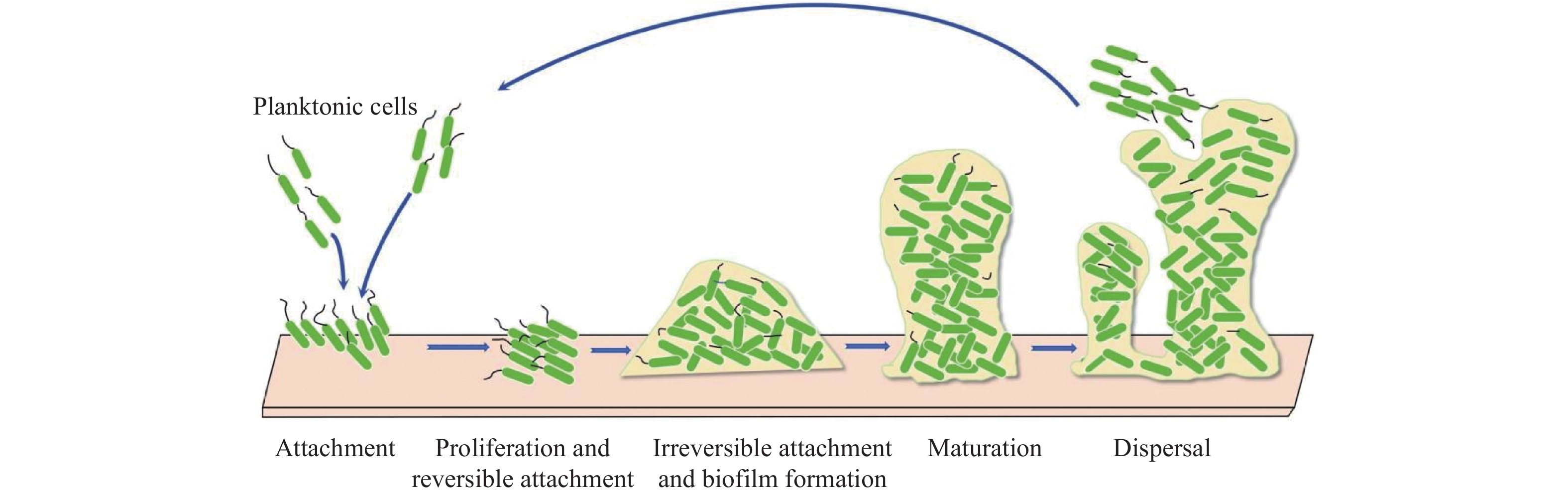
摘要: 大肠杆菌是一种适应性很强的条件致病菌,可以在植入物表面形成生物膜并产生持久细胞,导致危及生命的感染,抗生素难以治疗。因此,急需1种有效的大肠杆菌生物膜抑制剂来应对公共健康威胁。吲哚是近年发现的大肠杆菌新型群体感应信号分子,在调控细菌生长及生物膜形成方面具有重要意义,是未来研究新型抗生物膜制剂的潜在靶标。综述大肠杆菌生物膜的形成、吲哚的微生物代谢及其调控大肠杆菌生物膜形成研究进展,以期为临床治疗及药物研发提供帮助。Abstract: Escherichia coli is a highly adaptable opportunistic pathogen bacterium that can form biofilms on the surface of implants and generates persistent cells, leading to life-threatening infections that are difficult to treat with antibiotics alone. Therefore, there is a need for an effective E.coli biofilm inhibitor to combat this public health threat.Indole is a novel quorum-sensing signaling molecule of E. coli discovered in recent years, which is of great significance in regulating bacterial growth and biofilm formation, and is a potential target for future research on new anti-biofilm preparations. This article reviews the research progress on the formation of Escherichia coli biofilms, the microbial metabolism of indole and its regulation of Escherichia coli biofilm formation, in order to provide information for clinical treatment and drug development.
-
Key words:
- Escherichia coli /
- Indole /
- Bacterial biofilm /
- Biomaterials /
- Implant infection
-
新型冠状病毒(2019n-CoV)核酸检测结果阳性是确诊新型冠状病毒感染的依据[1]。本研究选取了2020年 2月至12月昆明市第三人民医院收治的59例确诊为新型冠状病毒感染者,探索咽拭子、粪便核酸阳性病例与咽拭子核酸阳性、粪便阴性病例之间是否存在不同的临床表现及2种样本的排毒规律,现报告如下。
1. 材料与方法
1.1 病例资料
选取2020年2至12月昆明市第三人民医院收治的59例新型冠状病毒感染者,诊断依据我国《新型冠状病毒肺炎诊疗方案(试行第七版)》[1]。59例均采集咽拭子、粪便标本。共采集咽拭子标本496份,其中阳性239份;共采集粪便标本209份,其中阳性68份。从首次采样至治愈出院,咽拭子及粪便均多次采集,直至连续3次检测阴性为止 。59例患者中,入院时处于潜伏期感染者13 例(22.03%),无症状感染者10例(16.95%),有症状感染者36例(61.02%)。59例中男35例(59.32%),女24例(40.68%),年龄(41.58±15.61)岁。2组一般情况(性别、年龄)比较差异均无统计学意义(P > 0.05),具有可比性。
1.2 研究方法
将新冠病例分为咽拭子、粪便核酸均阳性组(粪便阳性组或阳性组)与咽拭子核酸阳性、粪便核酸阴性组(粪便阴性或阴性组)2组。2组进行临床表现、肝功(r-谷氨酰氨基转移酶、门冬氨酸氨基转移酶、丙氨酸氨基转移酶)、T淋巴细胞(CD3+、CD4+、CD8+)、肾功能(尿素氮、肌酐、尿酸)、心肌酶(乳酸脱氢酶、激酸激酶、肌红蛋白)、血常规(白细胞、淋巴细胞计数、中性粒细胞计数、嗜酸性粒细胞计数)、超敏C反应蛋白(CRP)、肺部CT等对比;同时观察2组核酸排毒规律。
1.3 检测方法
实时荧光RT-PCR检测新型冠状病毒核酸。
1.4 统计学处理
实验数据用Excel软件对收集的数据进行整理,采用 SPSS 20.0 软件进行统计分析。计量资料不服从正态分布用中位数和四分位数[M(QR)]描述,两组组间比较采用非参数检验Wilcoxon 秩和检验;计数资料用例数和构成比[n(%)]描述,组间比较采用χ2检验,检验水准α = 0.05。
2. 结果
2.1 咽拭子、粪便核酸均阳性组与咽拭子阳性、粪便阴性组临床表现比较
59例患者均多次采集咽拭子及粪便标本,其中咽拭子及粪便核酸均阳性患者33例,咽拭子阳性、粪便阴性26例。胸部CT提示肺炎病例数,阳性组28例(84.85%),阴性组16例(61.54%),两组差异有统计学意义(P < 0.05);其余临床表现对比差异无统计学差异(P > 0.05),见表1。
表 1 粪便新冠核酸阳性病例与阴性病例临床表现比较[n(%)] (1)Table 1. Comparison of clinical manifestations between fecal COVID-19 positive cases and negative cases [n(%)] (1)粪便 发热 畏寒 咽痛 咽干 全身酸痛 阳性组 16(48.48) 2(6.06) 9(27.27) 8(24.24) 6(18.18) 阴性组 13(50.00) 3(11.54) 8(30.77) 3(11.54) 6(23.08) χ2 0.01 0.56 0.09 1.54 0.22 P 0.91 0.65 0.77 0.21 0.64 n = 59。 表 1 粪便新冠核酸阳性病例与阴性病例临床表现比较[n(%)] (2)Table 1. Comparison of clinical manifestations between fecal COVID-19 positive cases and negative cases [n(%)] (2)粪便 咯痰 咳嗽 气促 SaO2(≤93%) CT示肺炎病例数 阳性组 12(36.36) 18(54.55) 5(15.15) 9(27.27) 28(84.85) 阴性组 8(30.77) 11(42.30) 6(23.08) 3(11.54) 16(61.54) χ2 0.20 0.87 0.60 2.22 4.17 P 0.65 0.35 0.44 0.14 0.04* n = 59,*P < 0.05。 2.2 2组的血常规、CRP、肝功、肾功、T淋巴细胞对比
将粪便阳性组与阴性组的血常规、CRP、肝功、肾功、T淋巴细胞的各项指标进行对比,显示差异无统计学意义(P > 0.05),见表2。
表 2 粪便新冠核酸阳性病例与阴性病例实验室检查结果比较n = 59 [M(QR)]Table 2. Comparison of laboratory test results between fecal COVID-19 positive cases and negative cases n = 59 [M(QR)]项目(正常范围) 粪便核酸结果 [M(QR)] Z P 白细胞 + 4.94(1.79) −0.61 0.54 (4~10)×109/L − 5.62(30.70) 淋巴细胞计数 + 1.46(0.88) −0.53 0.60 (0.8~4)×109/L − 1.65(1.18) 中性粒细胞计数计数 + 3.09(2.33) −0.46 0.65 (2~7)×109/L − 3.09(2.75) 嗜酸性粒细胞计数 + 0.06(0.11) −0.38 0.71 (0.02~0.5) − 0.05(0.10) 血小板(×109/L) + 250(121.50) −1.79 0.07 (100~300)×109/L − 207(83) CRP + 1.30(4.87) −0.04 0.97 (0~6)mg/L − 1.35(12.04) CD4+计数 + 414.50(545.75) −1.58 0.11 (706~1125)个/μL − 495(531.75) CD3+计数 + 1004(640) −0.12 0.91 (1027~2086)个/μL − 1100(920.75) CD8+计数 + 371(306) −0.57 0.57 (323~836)个/μL − 382(340) 尿素氮 + 3.50(1.40) −1.72 0.09 (1.43~7.14) mmol/L − 3.10(1.27) 肌酐 + 63.10(28.40) −0.40 0.69 (35~97) μmol/L − 63.50(25.40) 尿酸 + 290.5(128) −0.91 0.37 (155~428) μmol/L − 267.350(143.53) r-谷氨酰氨基转移酶 + 25.20(20.90) −0.55 0.58 (7~50) U/L − 28.95(27.20) 丙氨酸氨基转移酶 + 22(13.50) −0.21 0.83 (5~40) U/L − 22.85(14.10) 门冬氨酸氨基转移酶 + 23(10.50) −0.54 0.59 (8~40) U/L − 23.5(10.50) 乳酸脱氢酶 + 176(26.50) −1.25 0.21 (109~245) U/L − 191(86.50) 肌酸激酶 + 79(47.10) −1.48 0.88 (26~174) U/L − 78.30(44) 肌红蛋白 + 17.44(10.02) −0.38 0.71 (0~68) ng/mL − 18.22(11.26) 2.3 咽拭子及粪便新型冠状病毒排毒规律
2.3.1 粪便阳性组咽拭子排毒规律
33例患者中:7例为潜伏期感染者,核酸首次阳性至出现症状时间为3~8 d(中位数为4 d),5例核酸持续阳性的时间为14~31 d(中位数为25 d),2例反复核酸阳性的时间为14 d和16 d;无症状感染者5例,2例咽拭子核酸持续阳性时间为4 d和16 d,2例反复咽拭子核酸阳性的时间4 d和22 d,1例核酸阳性1次。有症状感染者21例,发病至首次咽拭子核酸阳性时间为1~21 d(中位数为5 d),8例核酸持续阳性的时间为4~20 d(中位数为8 d);10例反复核酸阳性的时间为8~26 d(中位数为15 d),2例核酸阳性仅1次。
2.3.2 粪便阴性组咽拭子排毒规律
26例患者中:潜伏期感染者6例,咽拭子核酸首次阳性至出现症状时间为4~10 d(中位数为5 d),2例核酸持续阳性为2 d、3 d,2例核酸反复阳性时间分别共20 d和25 d,仅一次核酸阳性2例;无症状感染者4例,2例咽拭子核酸阳性时间为5 d和9 d,仅一次核酸阳性2例;有症状感染者16例,发病至首次咽拭子核酸阳性时间为1~16 d(中位数为5.5 d),3例核酸持续阳性的时间为2~7 d(中位数为6 d),10例反复核酸阳性时间共7~25 d(中位数为9 d),仅一次核酸阳性3例。
2组的咽拭子核酸阳性持续时间差异有统计学意义(P < 0.05),阳性组比阴性阳组持续排毒的时间长;2组的咽拭子核酸反复阳性持续时间差异无统计学意义(P > 0.05)。
2.3.3 新型冠状病毒感染者粪便排毒规律
33例粪便核酸阳性患者中:潜伏期感染者7例,4例粪便核酸持续阳性的时间3~14 d(中位数为4 d);咽拭子阳性后1~28 d粪便核酸阳性(中位数6 d)。无症状感染者5例,4例粪便核酸持续阳性的时间3~17 d(中位数为8 d);咽拭子阳性后5~20 d粪便阳性。有症状感染者21例,发病至首次粪便核酸阳性时间为4~29 d(中位数为15 d),10例粪便核酸持续阳性的时间为2~15 d(中位数为4.5 d);咽拭子阳性后4~29 d粪便阳性(中位数15 d)。
粪便阳性组间有症状感染者,首次咽拭子核酸阳性时间与粪便首次核酸阳性时间比较差异有统计学意义 (Z = -3.796,P < 0.01)。
2.3.4 临床分型
粪便阳性组潜伏期感染者、有症状感染者、无症状感染者分别占比为21.15%、63.6%、15.2%,阴性组中分别占比为23.1%、61.5%、15.4%;2组潜伏期感染者、有症状感染者、无症状感染者数量进行比较 χ2 = 0.157,P > 0.05差异无统计学意义。
临床分型情况:本组病例共59例,无症状感染者共9例(15.25%),轻型共10例(16.99%),普通型共28例(47.46%),重型共12例(20.34%)。粪便阳性组中无症状感染者5例(15.15%),无轻型病例,普通型19例(57.58%)、重型9例(27.27%);粪便阴性组中无症状感染者4例(15.39%)、轻型10例(38.46%)、普通型9例(34.62%)、重型3例(11.54%)。潜伏期感染者共13例(20.03%),其中出院诊断轻症3例占23.08%(阴性组3例),普通型10例占76.92%(阳性组7例,阴性组3例)。2组的轻症病例数差异有统计学意义(P < 0.01),阴性组的轻症病例多于阳性组;2组在无症状感染者、普通型、重症的病例数比较差异无统计学意义(P > 0.05)。
潜伏期感染者及无症状感染者,多因同伴发病或接触过患者来就诊。
2.3.5 转归
2组病例均使用干扰素雾化+洛匹那韦/利托那韦抗病毒治疗7~14 d,重症患者加用40~80 mg甲基强的松龙+静脉注射免疫球蛋白10~15 g 3 d。治疗后3~4周均痊愈。阳性组呼吸道最长排毒时间39 d、粪便最长排毒时间37 d,阴性组呼吸道最长排毒时间39 d。
3. 讨论
新型冠状病毒肺炎即COVID-19 (Coronavirus disease 2019)疫情自2019年底爆发以来,到2020年12月全球确诊病例超过580万例,死亡病例超过35万例,病死率约6.7%[1]。对新型冠状病毒特性及其临床特征的认识也不断增加。
新型冠状病毒(2019 novel coronavirus,2019-nCoV)属于β属的新型冠状病毒。传染源主要为新型冠状病毒感染者、无症状感染者,经呼吸道飞沫和密切接触传播是主要的传播途径,在相对封闭的环境中长时间暴露于高浓度气溶胶情况下存在经气溶胶传播的可能。在粪便中亦可分离到新型冠状病毒。人群普遍易感[2]。部分患者在呼吸道标本核酸检测阴性后,其粪便病原核酸检测仍可阳性[3]。
潜伏期1~14 d,多为3~7 d。可有发热、咽痛、咽干、咳嗽,可伴流涕、肌肉酸痛等轻症,有或无肺炎表现,部分患者无症状。重症患者多在发病1周后出现呼吸困难和/或低氧血症,进展为急性呼吸窘迫综合征、脓毒症休克、酸碱失衡、多器官功能衰竭等。胸部影像学早期呈现多发小斑片影及间质改变,以肺外带明显,进而发展为双肺多发磨玻璃影、浸润影,严重者可出现肺实变。发病早期外周血白细胞总数正常或减少,淋巴细胞计数减少,部分患者可出现肝酶、乳酸脱氢酶(LDH)、肌红蛋白增高;多数患者C反应蛋白(CRP)和血沉升高,降钙素原正常。咽拭子(或痰、肺泡灌洗液)、粪便、血液等标本经实时荧光RT-PCR或(或NGS)检测出新型冠状病毒核酸可确诊[2]。
本研究中,新型冠状病毒无症状感染者占15.25%,轻型占16.99%,普通型占47.46%,重型占20.34%,以普通型常见,重型占比不高。粪便阳性组与粪便阴性组比较有以下特点:(1)新型冠状病毒感染者常见症状发热、咽痛、咽干、全身酸痛、咳嗽、咯痰,气促、血氧饱和度低于93%等的例数,两组比较均无差异(P > 0.05);(2)入院时处于潜伏期感染者阳性组占21.21%、阴性组占23.08%,阳性组全部发展为普通型,阴性组发展为轻型、普通型各占一半,均无发展为重型及以上的病例;无症状感染者在阳性组占15.15%、阴性组占15.39%,多因同伴发病或接触过患者而就诊;有症状感染者在阳性组占63.64%(27.27%为重型、57.58%为普通型,无轻型),阴性组61.54%(11.54%为重症、34.62%为普通型、38.46%为轻型)。阴性组的轻症病例多于阳性组(P < 0.05),2组无症状感染者、普通型、重症的病例数比较无差异(P > 0.05);(3)肺部CT提示阳性组(84.85%)较阴性组(61.54%)更易出现肺炎(P < 0.05),与临床分型结果相符合;(4)粪便阳性组有症状感染者首次核酸阳性时间咽拭子比粪便出现得早(P < 0.01)[4];(5)2组的潜伏期感染者、有症状感染者、无症状感染者数量无明显差异 (χ2 = 0.157,P > 0.05);(6)在血常规、CRP、肝功、肾功、T淋巴细胞亚群等2组比较也无差异(P > 0.05);(7)2组的潜伏期、无症状携带状态、发病状态均存在排毒情况;(8)阳性组咽拭子排毒时间长于阴性组(P < 0.05)。2组咽拭子、粪便最长排毒时间均接近40 d;(9)潜伏期就诊的患者无1例发展成为重症,阳性组比阴性组更易进展为普通型;(10)2组的咽拭子核酸反复阳性时间差异无统计学意义(P > 0.05);(11)本组病例中粪便核酸检出时间基本落后于咽拭子,可能与检测技术有关,也可能消化道病原体排除时间就是落后于呼吸道。
综上所述,新型冠状病毒感染人体后,出现无症状携带者(隐性感染)及有临床症状(显性感染)两类患者。由于就诊时间的不一致,本组病例中有潜伏期感染者、轻症患者、普通型患者、重症患者几种临床类型。以普通型常见,重型占比不高,且由于发现及时,潜伏期感染患者得到及时救治而未发展为重症,早期诊断、早期干预治疗可减少向重症的发展。本组病例无论阳性组还是阴性组临床表现与指南中的表现无差异,没有特殊表现。粪便阴性组的轻症病例更多,但两组无症状感染者、普通型、重症的病例数却无差异,阴性组在肝、肾功,心肌酶,炎症指标方面病毒造成的机体损害不弱于阳性组,也需积极救治。阳性组肺部炎症病例明显多于阴性组,说明2个系统同时排毒可能存在体内的病原体更多,主要影响呼吸系统引起肺炎;咽拭子及粪便均能检出病原体的病例,除了说明排毒量多外,排毒时间也比粪便阴性组更长,作为传染源应该被严格管控。在临床表现全部消失后,仍然有排毒现象,两组咽拭子最长的检出时间相当(39 d),粪便最长检出时间(37 d)也与咽拭子的时间相当。宋旭辉等报道1例粪便排毒50 d[5],丛晓东等报道呼吸道排毒最长12周[6],陆晓东等报道粪便排毒时间长于咽拭子[4]。说明该病毒有长期被人体携带的特点,患者痊愈后仍然是防控工作的重点监管人群,而无症状感染者人群是防控中最大的难点。核酸反复出现阳性的问题不少见[7-8],排毒没有规律,所受的影响因素比较复杂[9],对防控极其不利,只有做好健康教育,嘱患者注意个人卫生及防护,一段时间内反复复查核酸,以免遗漏造成播散。呼吸道作为最早被侵犯的系统,病原体繁殖后首先排毒,故咽拭子较粪便中检出病原体的时间更早,咽拭子作为筛查新冠的手段更有意义。本组病例中粪便核酸检出时间落后于咽拭子,可能与检测技术有关,也可能因呼吸道为首先被感染的系统而先排病毒,消化道感染后于呼吸道,故排毒落后于呼吸道。可能因本组病例数有限,本组病例新冠病毒排毒欠规律性,也可能新冠病毒排毒就是无统一规律的,而且部分病例排毒时间较长,故需逐渐建立一套完善的病例管理方案,定期、定时限检测排毒情况及病毒变异情况。
-
[1] Li S,Duan W,Lei Y,et al. Effects of lipid emulsions on the formation of Escherichia coli-Candida albicans mixed-species biofilms on PVC[J]. Sci Rep,2021,11(1):16929. doi: 10.1038/s41598-021-96385-6 [2] Mei J,Xu D,Wang L,et al. Biofilm microenvironment-responsive self-assembly nanoreactors for all-stage biofilm associated Infection through bacterial cuproptosis-like death and macrophage re-rousing[J]. Adv Mater,2023,35(36):e2303432. doi: 10.1002/adma.202303432 [3] Li Q,Liu Q,Wang Z,et al. Biofilm homeostasis Interference therapy via 1O2-Sensitized hyperthermia and Immune microenvironment re-rousing for biofilm-associated Infections elimination[J]. Small,2023,19(22):e2300592. doi: 10.1002/smll.202300592 [4] Xu Q,Chen S,Jiang L,et al. Sonocatalytic hydrogen/hole-combined therapy for anti-biofilm and infected diabetic wound healing[J]. Natl Sci Rev,2023,10(5):nwad063. doi: 10.1093/nsr/nwad063 [5] 杨政鸿,何大千,宁明杰,等. 不同3D打印精度制作的生物材料表面形貌对表皮葡萄球菌生物膜形成影响[J]. 昆明医科大学学报,2022,43(2):12-17. doi: 10.12259/j.issn.2095-610X.S20220228 [6] 张国婧,万子琳,王小燕,等. 表皮葡萄球菌胞间黏附素基因操纵子对细菌与真菌混合生物膜相关炎症作用影响的体内研究[J]. 中国修复重建外科杂志,2021,35(10):1328-1335. [7] Lei Y,Xu Y,Jing P,et al. The effects of TGF-β1 on staphylococcus epidermidis biofilm formation in a tree shrew biomaterial-centered infection model[J]. Ann Transl Med,2021,9(1):57. doi: 10.21037/atm-20-4526 [8] 李民杰. 大肠杆菌运动调控在生物材料植入感染中的作用研究[D]. 昆明: 昆明医科大学, 2021. [9] Wang X,Zhang J,Chen W,et al. Study on the effects of estradiol in staphylococcus epidermidis device-related capsule formation[J]. Aesthetic Plast Surg,2020,44(2):558-569. doi: 10.1007/s00266-019-01567-3 [10] Karygianni L,Ren Z,Koo H,et al. Biofilm matrixome: Extracellular components in structured microbial communities[J]. Trends Microbiol,2020,28(8):668-681. doi: 10.1016/j.tim.2020.03.016 [11] Rosman C W K,van Dijl J M,Sjollema J. Interactions between the foreign body reaction and Staphylococcus aureus biomaterial-associated infection. Winning strategies in the derby on biomaterial implant surfaces[J]. Crit Rev Microbiol,2022,48(5):624-640. doi: 10.1080/1040841X.2021.2011132 [12] Zarkan A,Liu J,Matuszewska M,et al. Local and universal action: The paradoxes of Indole signalling in bacteria[J]. Trends Microbiol,2020,28(7):566-577. doi: 10.1016/j.tim.2020.02.007 [13] Bjarnsholt T,Buhlin K,Dufrêne Y F,et al. Biofilm formation-what we can learn from recent developments[J]. J Intern Med,2018,284(4):332-345. doi: 10.1111/joim.12782 [14] Arciola C R,Campoccia D,Montanaro L. Implantinfections: adhesion,biofilm formation and immune evasion[J]. Nat Rev Microbiol,2018,16(7):397-409. doi: 10.1038/s41579-018-0019-y [15] 羊扬,刘云,张信军,等. 大肠杆菌群体感应系统的研究进展[J]. 中国兽医学报,2018,38(8):1624-1631. doi: 10.16303/j.cnki.1005-4545.2018.08.27 [16] Flemming H C,Wuertz S. Bacteria and archaea on earth and theirabundance in biofilms[J]. Nat Rev Microbiol,2019,17(4):247-260. doi: 10.1038/s41579-019-0158-9 [17] 吴丽娜,董鹏程,张一敏,等. 大肠杆菌生物膜形成特性及控制措施的研究进展[J]. 食品科学,2019,40(15):307-313. doi: 10.7506/spkx1002-6630-20180910-100 [18] Berne C,Ellison C K,Ducret A,et al. Bacterial adhesion at the single-cell level[J]. Nat Rev Microbiol,2018,16(10):616-627. doi: 10.1038/s41579-018-0057-5 [19] Arnaouteli S,Bamford N C,Stanley-Wall N R,et al. Bacillus subtilis biofilm formation and social interactions[J]. Nat Rev Microbiol,2021,19(9):600-614. doi: 10.1038/s41579-021-00540-9 [20] Filipović U,Dahmane R G,Ghannouchi S,et al. Bacterial adhesion on orthopedic implants[J]. Adv Colloid Interface Sci,2020,283:102228. doi: 10.1016/j.cis.2020.102228 [21] Yan J,Bassler B L. Surviving as a community: Antibiotic tolerance and persistence in bacterial biofilms[J]. Cell Host Microbe,2019,26(1):15-21. doi: 10.1016/j.chom.2019.06.002 [22] Yin W,Wang Y,Liu L,et al. Biofilms: The microbial "protective clothing" in extreme environments[J]. Int J Mol Sci,2019,20(14):3423. doi: 10.3390/ijms20143423 [23] Roy R,Tiwari M,Donelli G,et al. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action[J]. Virulence,2018,9(1):522-554. doi: 10.1080/21505594.2017.1313372 [24] Del Pozo J L. Biofilm-related disease[J]. Expert Rev Anti Infect Ther,2018,16(1):51-65. doi: 10.1080/14787210.2018.1417036 [25] Balzan S,de Almeida Quadros C,de Cleva R,et al. Bacterial translocation: Overview of mechanisms and clinical impact[J]. J Gastroenterol Hepatol,2007,22(4):464-471. doi: 10.1111/j.1440-1746.2007.04933.x [26] Krawczyk B,Śledzińska A,Szemiako K,et al. Characterisation of Escherichia coli isolates from the blood of haematological adult patients with bacteraemia: Translocation from gut to blood requires the cooperation of multiple virulence factors[J]. Eur J Clin Microbiol Infect Dis,2015,34(6):1135-1143. doi: 10.1007/s10096-015-2331-z [27] Azimi S,Klementiev A D,Whiteley M,et al. Bacterial quorum sensing during Infection[J]. Annu Rev Microbiol,2020,74:201-219. doi: 10.1146/annurev-micro-032020-093845 [28] Coquant G,Grill J P,Seksik P. Impactof N-acyl-homoserine lactones,quorum sensing molecules,on gut Immunity[J]. Front Immunol,2020,11:1827. doi: 10.3389/fimmu.2020.01827 [29] Ahmed U K B,Ballard J D. Autoinducing peptide-based quorum signaling systems in clostridioides difficile[J]. Curr Opin Microbiol,2022,65:81-86. doi: 10.1016/j.mib.2021.10.017 [30] Yi L,Dong X,Grenier D,et al. Research progress of bacterialquorum sensing receptors: Classification,structure,function and characteristics[J]. Sci Total Environ,2021,763:143031. [31] Khera R,Mehdipour A R,Bolla J R,et al. Cryo-EM structures of pentameric autoinducer-2 exporter from Escherichia coli reveal its transport mechanism[J]. EMBO J,2022,41(18):e109990. doi: 10.15252/embj.2021109990 [32] Huang S,Liu X,Yang W,et al. Insights into adaptive mechanisms of extreme acidophiles[J]. Msystems,2022,7(2):e0149121. doi: 10.1128/msystems.01491-21 [33] Kim C S,Gatsios A,Cuesta S,et al. Characterization of autoinducer-3 structure and biosynthesis in E. coli[J]. ACS Cent Sci,2020,6(2):197-206. doi: 10.1021/acscentsci.9b01076 [34] Cui B,Chen X,Guo Q,et al. The cell-cell communication signal indole controls the physiology and interspecies communication of acinetobacter baumannii[J]. Microbiol Spectr,2022,10(4):e0102722. doi: 10.1128/spectrum.01027-22 [35] Wu S,Liu J,Liu C,et al. Quorum sensing for population-level control of bacteria and potential therapeutic applications[J]. Cell Mol Life Sci,2020,77(7):1319-1343. doi: 10.1007/s00018-019-03326-8 [36] Yaikhan T,Chuerboon M,Tippayatham N,et al. Indole and derivatives modulate biofilm formation and antibiotic tolerance of klebsiella pneumoniae[J]. Indian J Microbiol,2019,59(4):460-467. doi: 10.1007/s12088-019-00830-0 [37] Rattanaphan P,Mittraparp-Arthorn P,Srinoun K,et al. Indole signaling decreases biofilm formation and related virulence of Listeria monocytogenes[J]. FEMS Microbiol Lett,2020,367(14):fnaa116. doi: 10.1093/femsle/fnaa116 [38] Li Y,Feng T,Wang Y. The role of bacterial signaling networks in antibiotics response and resistance regulation[J]. Mar Life Sci Technol,2022,4(2):163-178. doi: 10.1007/s42995-022-00126-1 [39] Ganin H,Kemper N,Meir S,et al. Indole derivatives maintain the status quo between beneficial biofilms and their plant hosts[J]. Mol Plant Microbe Interact,2019,32(8):1013-1025. [40] Denamur E,Clermont O,Bonacorsi S,et al. The population genetics of pathogenic Escherichia coli[J]. Nat Rev Microbiol,2021,19(1):37-54. doi: 10.1038/s41579-020-0416-x [41] Gorelika O,Rogada A,Holoidovskyb L,et al. Meijlerb. Indole intercepts the communication between enteropathogenic E. coli and vibrio cholerae[J]. Gut Microbes,2022,14(1):2138677. doi: 10.1080/19490976.2022.2138677 [42] 韩茵,孙苗苗,王建平,等. 吲哚作为细菌细胞间信号分子的研究进展[J]. 微生物学通报,2015,42(4):736-748. doi: 10.13344/j.microbiol.china.140629 [43] Wang J,Zhang C,Childers W S. A biosensor for detection of Indole metabolites[J]. ACS Synth Biol,2021,10(7):1605-1614. doi: 10.1021/acssynbio.1c00090 [44] Fiore A,Murray P J. Tryptophan and indole metabolism in immune regulation[J]. Curr Opin Immunol,2021,70:7-14. doi: 10.1016/j.coi.2020.12.001 [45] 杨刚,张云露,李思明,等. 色氨酸对肠屏障免疫的调控作用研究进展[J]. 中国畜牧杂志,2021,57(4):6-10+16. doi: 10.19556/j.0258-7033.20200517-03 [46] Kumar A,Russell R M,Hoskan M A,et al. Indole sensing regulator (IsrR) promotes virulence gene[J]. mBio,2022,13(4):e0193922. doi: 10.1128/mbio.01939-22 [47] Han T H,Lee J H,Cho M H,et al. Environmental factors affecting indole production in Escherichia coli[J]. Res Microbiol,2011,162(2):108-116. doi: 10.1016/j.resmic.2010.11.005 [48] Li G,Young K D. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan[J]. Microbiology (Reading),2013,159(2):402-410. [49] Boon N,Kaur M,Aziz A,et al. The signaling molecule Indole Inhibits Induction of the AR2 acid resistance system in Escherichia coli[J]. Front Microbiol,2020,11:474. doi: 10.3389/fmicb.2020.00474 [50] Kumar A,SperandioV. Indole signaling at the host-microbiota-pathogen Interface[J]. mBio,2019,10(3):e01031-19. [51] Kim J,Shin B,Park C,et al. Indole-Induced activities of β-Lactamase and efflux pump confer ampicillin resistance in pseudomonas putida KT2440[J]. Front Microbiol,2017,8:433. [52] Cheng C,Yan X,Liu B,et al. SdiA enhanced the drug resistance of cronobacter sakazakii and suppressed Its motility,adhesion and biofilm formation[J]. Front Microbiol,2022,13:901912. doi: 10.3389/fmicb.2022.901912 [53] Mayer C,Borges A,Flament-Simon S C,et al. Quorum sensing architecture network in Escherichia coli virulence and pathogenesis[J]. FEMS Microbiol Rev,2023,47(4):fuad031. doi: 10.1093/femsre/fuad031 [54] Xuan G,Dou Q,Kong J,et al. Pseudomonas aeruginosa resists phage Infection via eavesdropping on Indole signaling[J]. Microbiol Spectr,2023,11(1):e0391122. doi: 10.1128/spectrum.03911-22 [55] Kim J,Park W. Indole: A signaling molecule or a mere metabolic byproduct that alters bacterial physiology at a high concentration?[J]. J Microbiology,2015,53(7):421-428. doi: 10.1007/s12275-015-5273-3 [56] Wang Y,Bian Z,Wang Y. Biofilm formation and inhibition mediated by bacterial quorum sensing[J]. Appl Microbiol Biotechnol,2022,106(19-20):6365-6381. doi: 10.1007/s00253-022-12150-3 [57] Liu W,Tang Q,Meng L,et al. Interbacterial chemical communication‐triggered nascent proteomics[J]. Angew Chem Int Ed Engl,2023,62(5):e202214010. doi: 10.1002/anie.202214010 [58] Sun F,Yuan Q,Wang Y,et al. Sub-minimum inhibitory concentration ceftazidime inhibits Escherichia coli biofilm formation by influencing the levels of the ibpA gene and extracellular indole[J]. Chemother,2020,32(1):7-14. doi: 10.1080/1120009X.2019.1678913 [59] Feng W,Zhang L,Yuan Q,et al. Effect of sub-minimal inhibitory concentration ceftazidime on the pathogenicity of uropathogenic Escherichia coli[J]. Microb Pathog,2021,151:104748. doi: 10.1016/j.micpath.2021.104748 [60] Lee J,Page R,García-Contreras R,et al. Structure and function of the E. coli protein YmgB: A protein critical for biofilm formation and acid-resistance[J]. J Mol Biol,2007,373(1):11-26. doi: 10.1016/j.jmb.2007.07.037 [61] Domka J,Lee J,Wood T K. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling[J]. Appl Environ Microbiol,2006,72(4):2449-2459. doi: 10.1128/AEM.72.4.2449-2459.2006 -






 下载:
下载:
 下载:
下载: