Bioinformatics-Based Analysis of the Roles of MX1,IFI44, and STAT1 in Lupus Nephritis
-
摘要:
目的 旨在筛选与LN相关的潜在生物标志物,以期用于早期诊断、病情监测和更精准的治疗方案制定。 方法 从基因表达谱数据库(gene expression omnibus,GEO)下载了GSE22221、GSE112943、GSE99967和GSE32591的基因表达数据。通过加权基因共表达网络分析(weighted gene co-expression network analysis,WGCNA)和微阵列数据的线性模型(linear models for microarray data,LIMMA),获得了交集基因。随后,利用基因本体论(gene ontology,GO)和京都基因与基因组百科全书(kyoto encyclopedia of genes and genomes,KEGG)对这些交集基因进行了生物功能和信号通路分析。接着,通过蛋白质-蛋白质相互作用(protein-protein interaction,PPI)网络分析、CytoHubba算法、支持向量机(support vector machine,SVM)和随机森林(random forest,RF)方法,筛选出了与LN高度相关的枢纽基因,进行了受试者工作特征曲线(receiver operating characteristic,ROC)分析,并利用GSE72798数据集对3个潜在的生物标志物进行了验证。 结果 WGCNA分析获得绿黄色模块(P = 7.4e−40)和青色模块(P = 1.5e−14);利用LIMMA法筛选到193个差异表达基因;共鉴定出113个LN相关的交集基因,GO和KEGG分析显示这些基因主要富集在病毒或细菌的防御、I型干扰素信号途径、中性粒细胞介导的免疫和Toll样受体信号等方面。通过CytoHubba、SVM和RF 3种方法筛选出MX1、IFI44和STAT1,其曲线下的面积(area under the curve,AUC)分别为0.874、0.879和0.833。验证数据集显示,MX1、IFI44和STAT1在LN患者中的表达显著高于健康人群(P < 0.001)。 结论 MX1、IFI44和STAT1在LN的发病机制中起到了关键作用,可能成为LN的重要生物标志物和未来的潜在治疗靶点。 Abstract:Objective To identify potential biomarkers associated with LN, with the goal of improving early diagnosis, disease monitoring, and the development of more precise treatment strategies. Methods Gene expression data were downloaded from the Gene Expression Omnibus (GEO) database for datasets GSE22221, GSE112943, GSE99967, and GSE32591. Intersecting genes were obtained through the application of weighted gene co-expression network analysis (WGCNA) and linear models for microarray data (LIMMA). Subsequently, biological function and pathway analyses were conducted on these intersecting genes using Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). Next, protein-protein interaction (PPI) network analysis was performed, and hub genes highly associated with LN were identified using the CytoHubba algorithm, support vector machine (SVM), and random forest (RF) methods. Receiver operating characteristic (ROC) analysis was performed, and three potential biomarkers were validated using the GSE72798 dataset. Results The green-yellow module (P = 7.4e−40) and the cyan module (P = 1.5e−14) were identified through WGCNA analysis. A total of 193 differentially expressed genes were identified using LIMMA, with 113 intersecting genes related to LN being identified. GO and KEGG analyses indicated that these genes were mainly enriched in viral or bacterial defense, type I interferon signaling pathway, neutrophil-mediated immunity, and Toll-like receptor signaling. MX1, IFI44, and STAT1 were identified as hub genes using CytoHubba, SVM, and RF methods, with AUC values of 0.874, 0.879, and 0.833, respectively. Validation using the GSE72798 dataset demonstrated that the expression of MX1, IFI44, and STAT1 was significantly higher in LN patients compared to healthy individuals (P < 0.001 for all). Conclusion MX1, IFI44, and STAT1 play crucial roles in the pathogenesis of LN and may serve as important biomarkers and potential therapeutic targets for LN. -
Key words:
- Lupus nephritis /
- Systemic lupus erythematosus /
- Biomarker /
- Hub genes /
- Type I interferon
-
系统性红斑狼疮(systemic lupus erythematosus,SLE)是1种由多种因素诱发的自身免疫性疾病,这些因素包括雌激素、紫外线、药物、污染、维生素D缺乏和病毒等[1]。该病可影响多个器官,包括关节、肺、皮肤和肾脏。在其并发症中,狼疮性肾炎(lupus nephritis ,LN)是最严重的,其病理特征为肾内淋巴组织形成、产生自身抗体和免疫复合物沉积。全球LN的发病率因地区和种族的不同而有所差异,亚洲的发病率和严重性均高于西方国家[2]。约60%的SLE患者在10 a内发展为LN,其中10%在5 a内进展为终末期肾病(end stage renal disease,ESRD),需要透析或移植[3]。LN是导致患者残疾和死亡的主要原因。
2021年1月,voclosporin的批准上市,结束了LN治疗数10 a无新药的局面[4]。在初始治疗阶段,低剂量环磷酰胺或霉酚酸酯(mycophenolate mofetil,MMF)被用作高剂量环磷酰胺的替代。尽管这些治疗减少了相关副作用,但它们并不比高剂量环磷酰胺更为有效[5]。
研究显示,LN诊断时如出现严重器官损害、高血压、白蛋白减少和肾功能下降,ESRD风险增加,这强调了早期诊断的重要性[3]。与无LN患者相比,伴有LN的狼疮患者死亡率显著增加,并且在增生性LN患者中,5 a内约有5%至25%的死亡直接归因于肾病。此外,10%至30%的LN患者会进展至需要肾脏替代治疗的肾衰竭。患有增生性LN(III、IV或III/IV + V类)的人群面临更高的肾脏替代治疗需求风险[6]。因此,早期诊断和治疗显著提高生存率。狼疮性肾炎的组织学分类指导治疗策略,旨在减少蛋白尿并维护肾功能。尽管肾活检是LN确诊的金标准,但由于其侵入性,使用受到限制。关于肾损伤与LN患者肾活检实际价值的争议仍然存在[7]。
随着表达谱芯片数据的兴起和生物信息学的进步,研究人员开始在基因水平上探讨疾病机制[8]。开发基于基因或蛋白的生物标志物以预测和监测LN肾损害对早期诊断和治疗至关重要。WGCNA是1种描述基因关联模式的系统方法,能够筛查与疾病相关的基因模块[9]。SVM是1种主要用于分类和回归的有监督学习模型,是深度学习之前的关键算法[10]。RF是1种由多决策树组成的分类器集合,通过所有树的投票决定输出类别,并在属性选择中引入随机性[11]。这些方法广泛应用于疾病基因研究。然而,利用这些方法筛选LN核心基因的研究仍然很少,大多数研究所选用的芯片数量有限且来源单一,导致筛选基因存在差异。
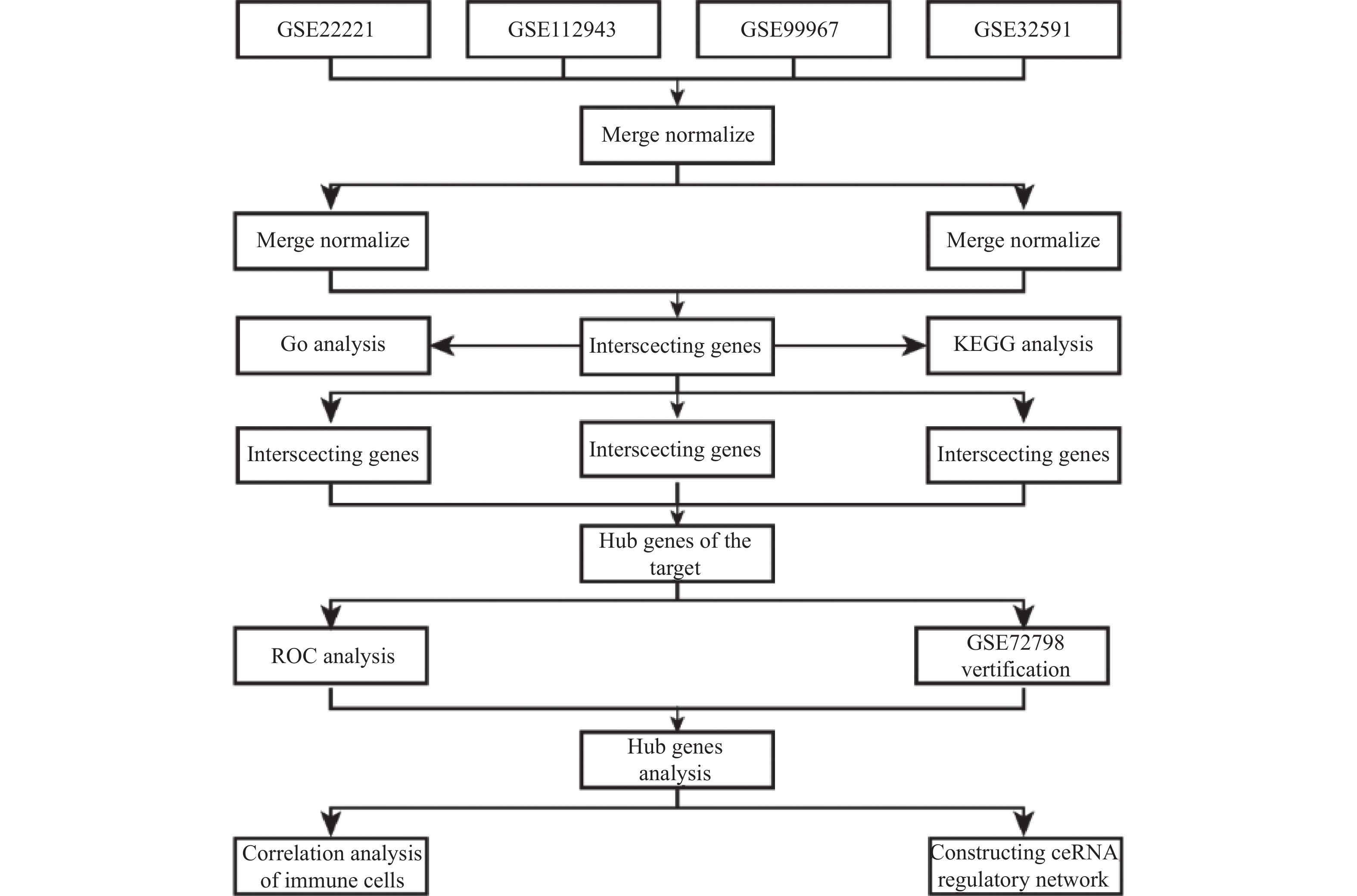
在本研究中,笔者选择了人类的肾和血液组织样本,采用了4个表达谱芯片并进行了批次校正。首先,笔者使用“limma”包确定了LN的差异表达基因(DEGs),并使用WGCNA构建基因共表达网络,找出与LN相关的基因模块。然后,将DEGs与基因模块交叉,并进行GO和KEGG分析。接着,结合Cytohubba插件、SVM和RF方法分析交集基因,并取这些方法得到基因的交集,确定目标核心基因。最后,利用ROC曲线和GSE72798数据集对核心基因进行了验证,并分析了它们与免疫细胞的相关性及在ceRNA调控网络中的作用,见图1。这些核心基因可能与LN的病理机制和进展紧密相关,对LN的早期诊断和治疗具有关键意义。
1. 资料与方法
1.1 数据来源
从GEO数据库(https://www.ncbi.nlm.nih.gov/geo/)提取以下基因芯片数据:GSE22221中,15份红斑狼疮患者血样(GSM2159869-GSM2159883)与25份健康者血样(GSM2159829-GSM2159853); GSE112943中,14份红斑狼疮患者肾脏组织样(GSM3091782、GSM3091791-GSM3091795、GSM3091783-GSM3091790)与7份健康者肾脏组织样(GSM3091775-GSM3091781); GSE99967中,29份红斑狼疮患者血样(如GSM2666720、GSM2666722-GSM2666725等)与17份健康者血样(GSM2666758-GSM2666774); GSE32591中,64份红斑狼疮患者肾脏组织样(GSM807842-GSM807873、GSM807889-GSM807920)与29份健康者肾脏组织样(GSM807874-GSM807888、GSM807921-GSM807934); GSE72798中,30份红斑狼疮患者血样(GSM1869396-GSM1869425)与10份健康者血样(GSM1869520-GSM1869529)。注释平台包括GPL10558、GPL21970、GPL14663和GPL570。训练组为GSE22221、GSE112943、GSE99967、GSE32591,验证组为GSE72798,见表1。
表 1 实验数据集信息Table 1. Experimental dataset information1.2 数据预处理
利用“strawberry-perl-5.32.0.1”将表达矩阵的探针信息映射至基因名。笔者用R-4.2.3对原始表达矩阵进行细致处理,通过“limma”包完成数据的规范化、标准化和归一化,并使用“sva”包的ComBat函数对训练组数据执行批次修正,同时对修正前后数据进行可视化。
1.3 加权共表达网络构建与分析
WGCNA方法分析基因表达数据,该方法在系统生物学中被广泛采用。用R语言的WGCNA包创建共表达网络,通过pickSoftThreshold函数确定最佳的软阈值参数。接着,建立加权邻接矩阵,并基于拓扑重叠矩阵(TOM)的分层聚类创建相关基因模块。
1.4 交集基因的筛选和功能富集分析
用R语言中的“limma”包,设置条件为|Log2FC|> 0.585 和P < 0.05,从LN和正常肾组织样本中挑选差异表达基因。通过“VennDiagram”包得到并可视化交集基因,这些交集基因是WGCNA中筛选模块与差异表达基因的共同致病相关基因。借助“clusterProfiler”、“limma”、“org.Hs.eg.db”、“enrichplot”及“pathview”包,对差异基因进行GO功能和KEGG信号通路富集分析。
1.5 Cytohubba筛选核心基因
将筛选出的交集基因导入 STRING 数据库构建 PPI网络,关键参数设置: Multiple proteins; Homo sapiens; highconfidence ( 0.700 ) ; hide disconnected nodes in thenetwork。使用 Cytoscape3.9.1的插件“Cytohubba”,获取MCC、DMNC、MNC、Degree、EcCentricity、BottleNeck、Closeness、Radiality、Betweenness和Stress打分最高的前50个基因。再从这些基因中找出交集,得到Cytohubba的核心基因。
1.6 支持向量机和随机森林筛选核心基因
RF和SVM算法在多学科交叉和机器学习的推动下,已成为基因表达谱数据分析的重要工具。利用R中的“glmnet”和“ggplot2”包,笔者对交集基因进行SVM分析并进行了可视化。准确性最高的点和误差最低的点所对应的基因数,均为SVM筛选LN特征基因的数量。同样地,使用“randomForest”和“ggplot2”包对交集基因进行了RF分析,并进行了可视化。决策树数量是决定模型表现的关键因素。通过交叉验证,笔者在误差曲线上找到了最小的交叉验证误差,并据此确定了最优决策树数量。此外,笔者还计算了基因的重要性得分。
1.7 获得目标核心基因
通过Cytohubba、SVM和RF方法分别筛选得到核心基因,利用R语言中的‘VennDiagram’包对这些核心基因进行交集分析和可视化,最终获得目标核心基因。
1.8 验证目标核心基因
本研究使用GSE72798数据集作为外部验证数据集,利用 R 语言的 “limma”、“ggpubr” 和“phteamap”包比较目标核心基因在在LN患者和健康人组织之间的表达差异分析。利用 R 语言的“pROC”包用以ROC分析并计算了AUC,以评价目标核心基因的预测效力。
1.9 统计学处理
所有的统计分析都使用R 4.2.3软件,采用Wilcox检验比较2组间差异,P < 0.05为差异有统计学意义。
2. 结果
2.1 联合芯片批次矫正
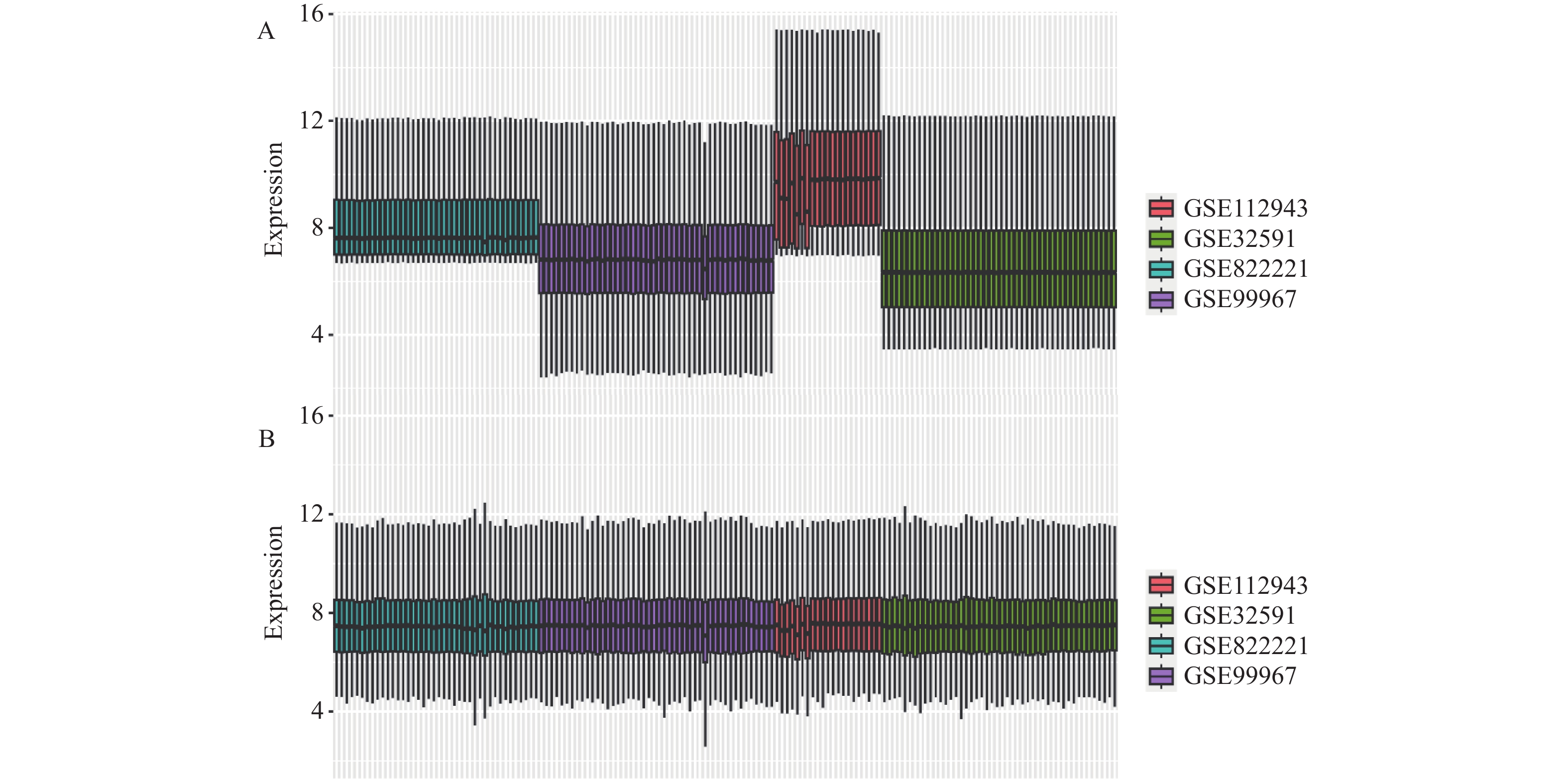
鉴于LN的复杂性和多样性,本研究致力于探索和识别其相关的生物标记物。由于相同组织不同批次的芯片以及不同组织来源的芯片在基因表达上都存在较大的差异,这种差异可能影响生物标记物的准确鉴定。因此,本研究的核心目标是通过多芯片批次矫正技术,消除这些差异,以获得更为稳定和准确的基因表达数据,见图2。在没有进行多芯片批次矫正的情况下,4个选定的表达谱芯片之间的基因表达差异明显。为了提高数据的稳定性和一致性,采用了多芯片批次矫正技术。矫正后,这4个芯片在基因表达量上的差异显著减少,数据呈现出1种归一化的分布特征,说明矫正技术有效地消除了不同批次和不同组织来源的芯片间的差异。通过这种方法,笔者期望在后续的生物信息学分析中获得更为准确和有力的支持,从而更有效地识别LN的潜在生物标记物。
2.2 共表达网络构建及重要模块的识别
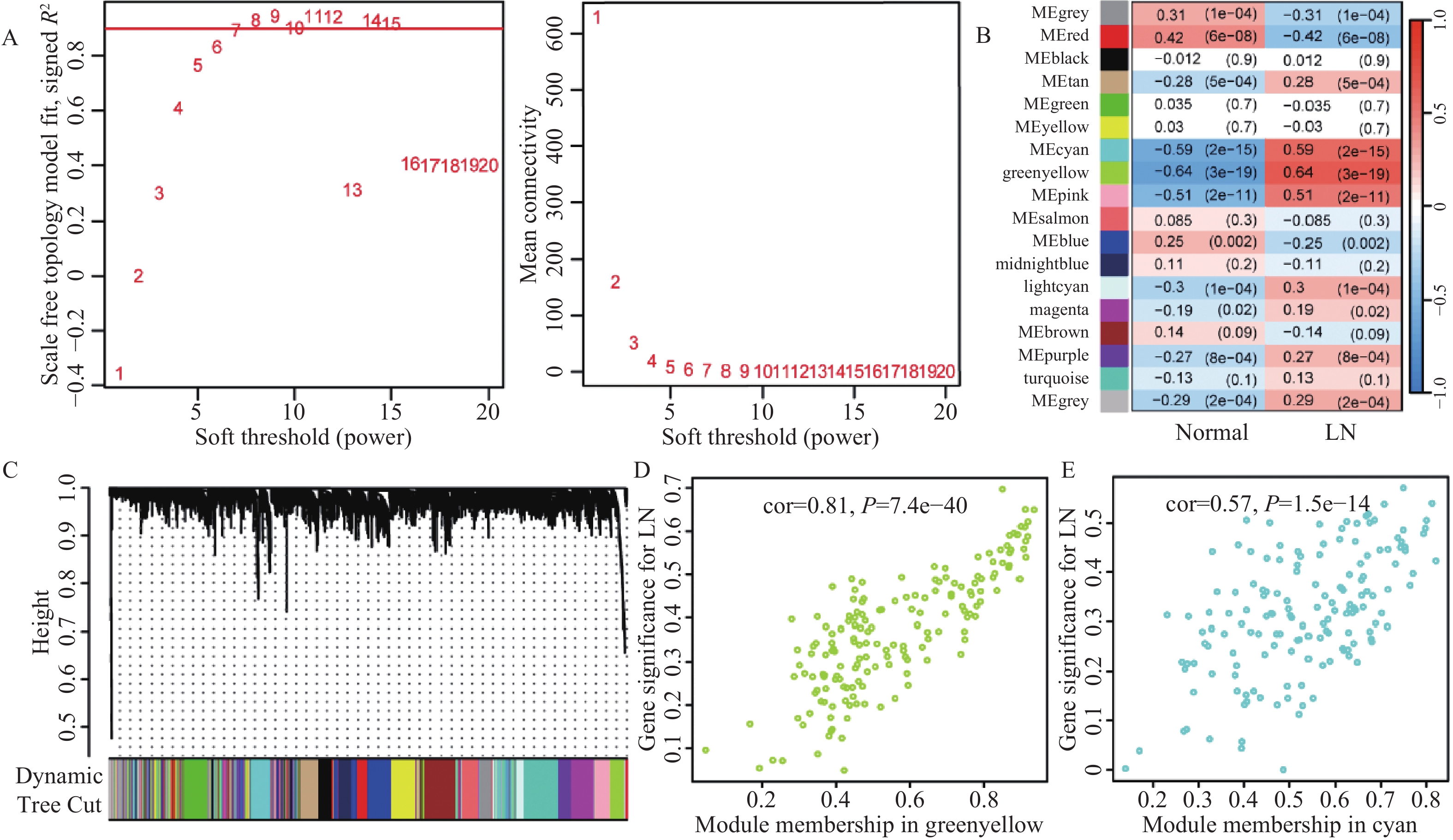
为了更深入地探索LN的生物学特性及其潜在生物标志物,笔者计划应用WGCNA技术,分析LN患者和正常对照组的芯片数据,识别与LN密切相关的基因模块。基于200个样本(包括122个LN患者样本和78个正常对照样本)的联合芯片数据集,构建了WGCNA网络。选择软阈值是关键步骤,因此,根据无尺度拟合指数R²首次达到0.9,确定最佳软阈值为β=7,见图3A。在这一软阈值下,网络的平均连接程度较高,确保在分析中捕获足够的信息量。进一步采用了模块合并策略,将模块相似性阈值(MEDissThres)设置为0.25,以确保合并距离较近且具有相似表达模式的模块。同时,设定每个模块至少包含50个基因,并设置剪切高度为0.25。经过这些步骤,共得到了18个与LN相关的基因模块,见图3B,3C。在随后的模块-特征关系分析中,热图显示了各个模块与LN的相关性。值得注意的是,绿黄色模块和青色模块与LN的相关性最为显著。具体而言,绿黄色模块的相关性为cor = 0.81(P = 7.4e−40);青色模块的相关性为cor = 0.57(P = 1.5e−14),见图3D。
2.3 LN差异表达基因的筛选
为了深入了解LN的分子机制,计划鉴定与LN相关的差异表达基因。在对联合基因芯片数据进行标准化处理后,成功鉴定出193个与LN相关的差异表达基因,其中183个基因呈现上调表达(占比为94.8%),而10个基因呈现下调表达(占比为5.2%),见表2。
表 2 正常组与LN患者差异基因Table 2. Differential genes between normal group and LN patients差异基因 上调基因 IFI27,IFI44L,IFI44,MX1,LTF,RSAD2,CD163,HERC5,PLSCR1,OAS3,IFIT3,OAS1,OAS2,IFI6,IFIT1,LY96,MX2,HERC6,CREG1,SIGLEC1,EIF2AK2,CEACAM8,C1QB,IFITM1,IFIH1,IFIT2,DDX60,DYSF,ELANE,DEFA4,ABCA1,XAF1,IFITM3,TRIM22,ISG15,LCN2,RNASE6,OLFM4,APOBEC3A,IFI35,RNASE1,VSIG4,MMP9,DHX58,RNASE2,TOP2A,TLR2,LYN,HCK,HBD,MMP8,IRF7,ISG20,ZCCHC2,OLR1,CDC20,GBP2,MS4A4A,DDX58,CCL2,RRM2,SPATS2L,LHFPL2,GBP1,CTSG,CEACAM1,STAT1,CD38,ENTPD1,UBE2J1,TCN2,CXCL10,RTP4,FCER1G,TYMS,B4GALT5,CCNA2,RP2,C3AR1,SAMD9,YIPF6,CARHSP1,TAP1,IFI30,LGALS3BP,TRIM21,FAR2,STIL,PARP12,MFGE8,RNASE3,H2AFZ,CEACAM6,LAP3,YWHAH,GMPR,RIN2,SCARB2,LMNB1,MPO,LCP1,BPGM,TLR4,PRC1,LY6E,CFD,MS4A6A,TPX2,XK,LRRC32,SH3GLB1,TGM2, CTSS,OASL,COL1A2,GINS2,ANP32B,ADAR,RAPGEF5,LYZ,MARCKS,HMGB2,ATP6V1D,SP100,FAM46C,CAMP,COL4A1,TMEM140,ANXA3,FCN1,NETO2,MYOF,EVI2A,FUT4,DNAJC13,CD36,IFI16,POU2AF1,SAMSN1,TCN1,MR1,MNDA,GIMAP4,PRCP,NFE2,CA1,STAT2,PPP1R3D,TACC3,SLPI,C1QA,S100A8,NMI,TLR1,NUSAP1,HLX,ARG1,TMOD1,REXO2,PLA2G4A,PTEN,TNFSF10,NPL,PLEK,GINS3,NOD2,LXN,DNAJC15,HIST1H2AI,MFSD1,DEGS1,FAM46A,CENPN,ITGB2,SCO2,PSMC2,COL15A1,CDC123,DHRS9,HK3,SERPINB1,NOL7,ITGAM 下调基因 CLIC5,ASB1,PTGDS,EPHX2,CDKN1C,TNNC2,KIR2DL3,FCER1A,TGFBR3,FOSB 2.4 交集基因的筛选及富集分析
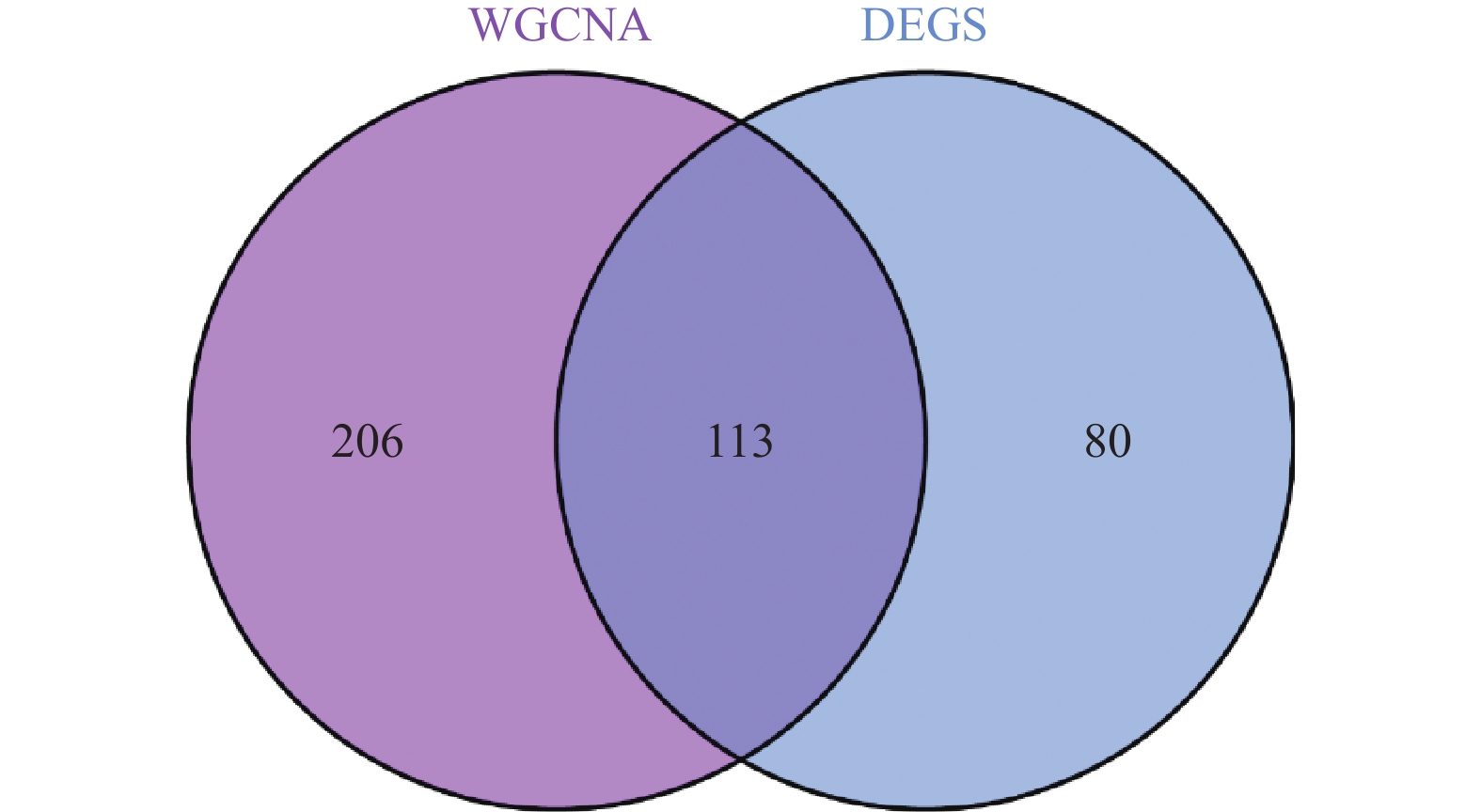
为了鉴定在LN中与差异表达基因(differentially expressed genes,DEGs)相关的关键模块基因,选取了绿黄色模块和青色模块的基因进行分析。与DEGs的交集分析鉴定出113个可能的LN相关的关键基因,见图4。利用R 4.2.3版本进行的功能富集分析表明:在生物过程(biological process,BP)中,这些基因主要与病毒的防御、I型干扰素信号、细菌的防御、细胞因子信号、模式识别受体信号、RIG-I信号、中性粒细胞介导的免疫、Toll样受体信号以及调节性T细胞的免疫等过程有关;在细胞组分(cellular component,CC)中,显著富集于特异性颗粒、分泌颗粒管腔、细胞质囊泡腔体、初级溶酶体和吞噬细胞囊泡;而在分子功能(molecular function,MF)中,它们主要涉及双链RNA的结合、DNA的结合、核酸酶活性、趋化因子受体的结合以及激酶的调节活性,见表3。此外,KEGG路径分析揭示了这些基因在甲型流感、COVID-19、NOD受体信号、RIG-I信号、金黄色葡萄球菌感染、SLE、IL-17信号、细胞膜DNA感应、补体途径、程序性坏死、Toll样受体信号以及中性粒细胞外陷阱形成等路径中可能发挥的重要作用,见表4。
表 3 交集基因GO富集分析Table 3. GO enrichment analysis of intersection genesONTOLOGY ID 描述 P BP GO:0051607 defense response to virus 0.000 GO:0060337 type I interferon signaling pathway 0.000 GO:0042742 defense response to bacterium 0.000 GO:0019221 cytokine-mediated signaling pathway 0.000 GO:0002221 pattern recognition receptor signaling pathway 0.000 GO:0039529 RIG-I signaling pathway 0.000 GO:0002446 neutrophil mediated immunity 0.000 GO:0002709 toll-like receptor signaling pathway 0.000 GO:0002709 regulation of T cell mediated immunity 0.002 CC GO:0042581 specific granule 0.000 GO:0034774 secretory granule lumen 0.000 GO:0060205 cytoplasmic vesicle lumen 0.000 GO:0005766 primary lysosome 0.000 GO:0045335 phagocytic vesicle 0.001 MF GO:0003725 double-stranded RNA binding 0.000 GO:0008301 DNA binding 0.006 GO:0004518 nuclease activity 0.008 GO:0042379 chemokine receptor binding 0.009 GO:0019207 kinase regulator activity 0.015 表 4 交集基因KEGG信号通路分析Table 4. KEGG pathway analysis of intersection genesID 描述 P hsa05164 Influenza A 0.000 hsa05171 COVID-19 0.000 hsa04621 NOD-like receptor signaling pathway 0.000 hsa04622 RIG-I-like receptor signaling pathway 0.000 hsa05150 Staphylococcus aureus infection 0.001 hsa05322 Systemic lupus erythematosus 0.002 hsa04657 IL-17 signaling pathway 0.004 hsa04623 Cytosolic DNA-sensing pathway 0.016 hsa04610 Complement signaling pathway 0.022 hsa04217 Necroptosis 0.025 hsa04620 Toll-like receptor signaling pathway 0.037 hsa04613 Neutrophil extracellular trap formation 0.044 2.5 枢纽基因的筛选
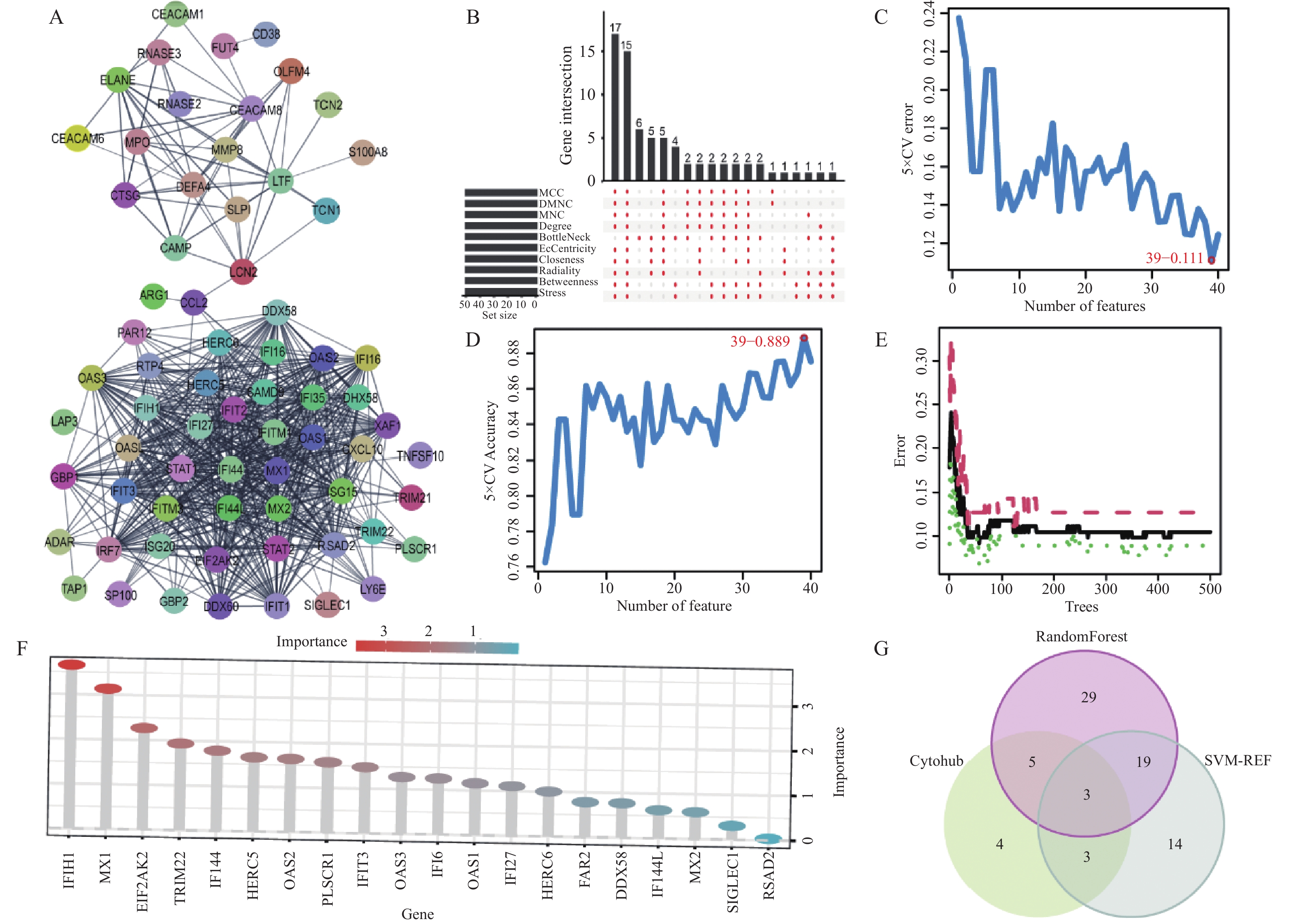
在无尺度网络中,大多数节点只有少量的连接,而少数节点(称为“枢纽节点”)有大量的连接。生物网络中的小部分基因(枢纽基因)可能在许多生物过程中起到非常重要的作用。为了深入探讨LN高度相关的枢纽基因,笔者从交集中挑选了113个基因并以此构建了PPI)网络。该网络涉及76个节点和615个互作关系。通过使用Cytoscape软件的cytoHubba插件,成功鉴定了以下15个网络中的核心基因:DDX60、IFI44、HERC5、HERC6、RTP4、DHX58、GBP1、IFI44L、IFIT1、IRF7、MX1、RSAD2、XAF1、STAT1和CXCL10。利用SVM方法,进一步从交集基因中确定了39个具有显著特征的基因:GINS3、LY6E、CDC20、MARCKS、ELANE、PRC1、PTGDS、MX1、ARG1、RSAD2、FAR2、TAP1、TPX2、LGALS3BP、TCN2、RNASE1、ADAR、SP100IFI6、SAMD9、ZCCHC2、CEACAM1、RTP4、MYOF、XAF1、STAT1、OAS2、IFITM3、IFI44、EIF2AK2、ANXA3、FUT4、IFIT2、TYMS、RAPGEF5、RNASE2、CTSG、SLPI和LAP3。此外,通过RF算法,鉴定了56个基因具有重要的诊断价值,包括:IFIH1、MX1、CDC20、RNASE1、LTF、EIF2AK2、RNASE2、TRIM22、IFI44、HMGB2、HERC5、OAS2、PLSCR1、TPX2、MPO、IFIT3、ELANE、GINS2、OAS3、IFI6、DEFA4、OAS1、IFI27、RRM2、HERC6、STIL、LXN、GBP1、GINS3、SCARB2、FAR2、DDX58、PRC1、TOP2A、RAPGEF5、ARG1、CEACAM1、TCN2、IFI44L、TGFBR3、MX2、IFITM1、NUSAP1、CDKN1C、OLR1、MMP8、MS4A4A、CTSG、RIN2、SP100、FAM46A、SLPI、STAT1、IRF7、SAMD9和TRIM21。
综合以上3种方法的结果,并通过韦恩图分析,确定了3个具有重要诊断潜力的LN相关生物标记物,分别是:IFI44、MX1和STAT1,见图5。
2.6 枢纽基因的验证分析
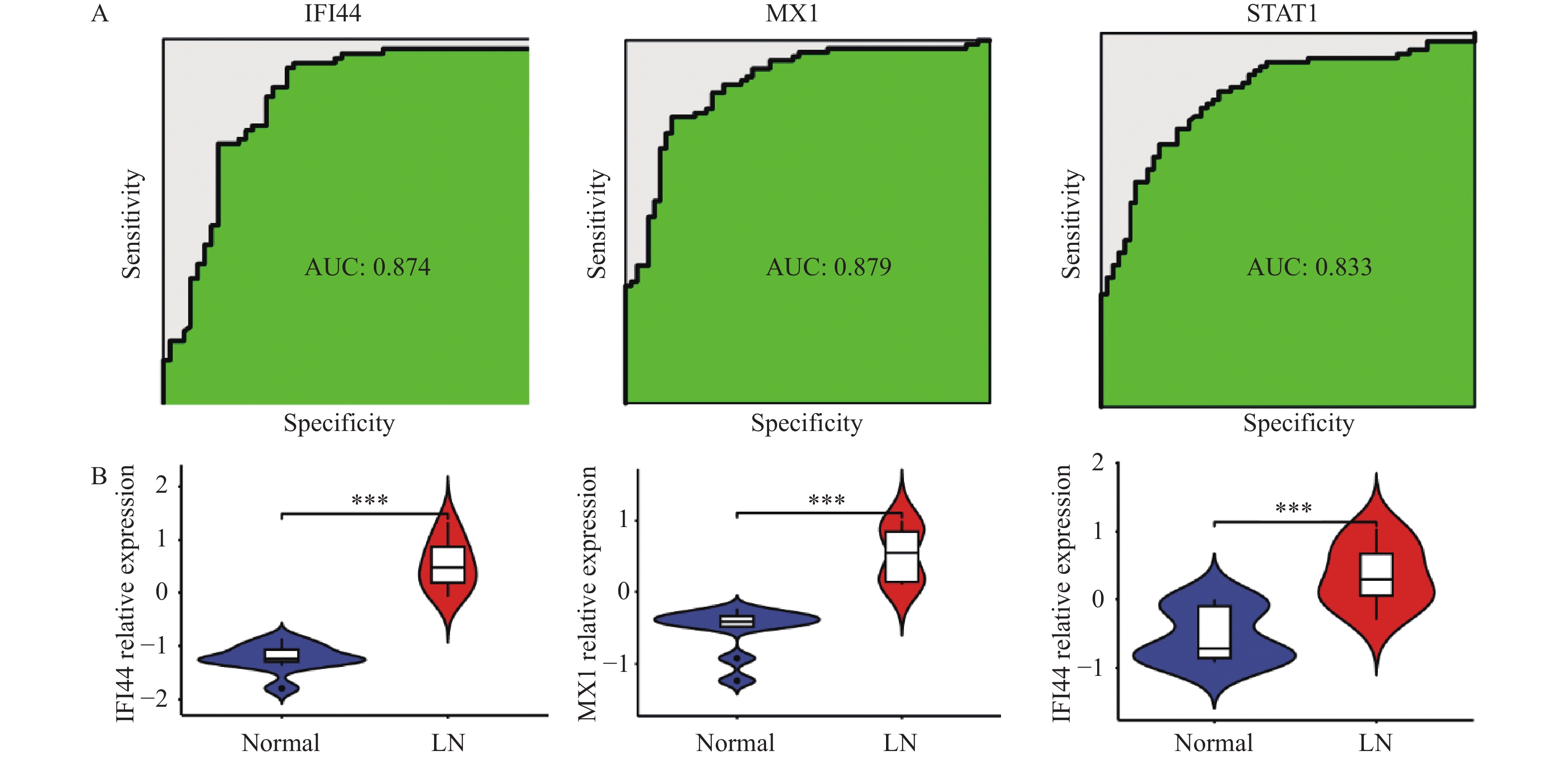
为了验证生物信息学方法所预测的3个关键基因IFI44、MX1和STAT1的准确性,首先通过接收者操作特性曲线(ROC)的分析,发现IFI44、MX1和STAT1在LN的诊断和预后上均表现出良好的鉴别能力,其AUC值分别为0.874、0.879和0.833,这表明这3个基因在LN中具有较高的诊断准确性。为进一步验证这些初步结果,采用了外部数据集GSE72798。结果显示,在LN患者与健康对照组之间,IFI44、MX1和STAT1在血液组织中的相对表达量有统计学意义,其中LN患者的表达显著高于健康人群(P < 0.001),见图6。
3. 讨论
3.1 I型干扰素信号通路在LN发病中的关键作用
LN是SLE的主要并发症,严重影响患者的生活质量和预期寿命。本研究结合多基因芯片分析、WGCNA与LIMMA技术,筛选出与LN相关的模块基因及差异表达基因,并进行了GO和KEGG功能富集分析。结果表明,相关通路主要涉及病毒感染、I型干扰素信号通路、细胞因子介导的信号传导及免疫反应等。研究表明,LN的发生可能与免疫复合物、病毒感染及干扰素介导的免疫反应密切相关。I型干扰素,特别是IFN-α和IFN-β,在LN的发病中起关键作用,模式识别受体感知病毒并激活IFN-I的产生,进一步加剧肾脏损伤。因此,干扰素信号通路在LN的病理机制中占据核心地位,验证了其作为潜在治疗靶点的价值。
3.2 核心基因MX1、IFI44和STAT1的作用机制
通过多种生物信息学方法,本研究鉴定了3个与LN密切相关的核心基因:MX1、IFI44和STAT1,这些基因在I型干扰素信号通路中发挥重要作用。
3.2.1 MX1的作用
MX1是由干扰素激活的先天免疫应答蛋白,主要通过抑制RNA病毒的复制发挥作用[17]。研究显示,MX1在LN的免疫反应和肾脏纤维化过程中起核心作用,且与干扰素信号途径密切相关[18-19]。抑制MX1可显著减轻肾纤维化[18],表明其在LN中的免疫调节作用。
3.2.2 IFI44的作用
IFI44是干扰素刺激基因,在免疫反应中具有重要作用[20]。研究发现,LN患者中IFI44的表达显著增高,可能通过调节先天免疫反应来参与LN的发病过程[21],表明IFI44可能成为LN的关键生物标志物。
3.2.3 STAT1的作用
STAT1在干扰素信号通路中扮演关键角色,并与免疫应答密切相关[22]。研究表明,STAT1的活化与LN的病理变化及疾病活跃度密切相关,尤其在巨噬细胞的聚集和肾功能损害中具有重要作用[23−25]。尽管STAT1在LN中的作用已得到初步验证,但其具体机制仍需进一步探索。
MX1、IFI44和STAT1作为干扰素响应基因,其上调表达表明I型干扰素信号通路的激活在LN中的关键作用。干扰素通过促进免疫细胞迁移、激活炎症因子及诱导细胞凋亡,可能导致肾脏的持续损伤和纤维化[26-27]。针对干扰素信号通路的干预可能为LN的治疗提供新的思路。
3.3 研究创新点及展望
本研究首次综合应用WGCNA、LIMMA、PPI、CytoHubba、SVM和RF等多种分析方法,成功鉴定出MX1、IFI44和STAT1这3个与LN密切相关的核心基因,并通过GO和KEGG功能富集分析验证了它们在I型干扰素信号通路中的关键作用。这些基因不仅为LN的诊断和预后提供了潜在的生物标志物,还为LN的靶向治疗提供了新的研究方向。未来的研究可进一步探索这些基因在LN中的具体机制,并验证其在临床应用中的可行性。
-
表 1 实验数据集信息
Table 1. Experimental dataset information
表 2 正常组与LN患者差异基因
Table 2. Differential genes between normal group and LN patients
差异基因 上调基因 IFI27,IFI44L,IFI44,MX1,LTF,RSAD2,CD163,HERC5,PLSCR1,OAS3,IFIT3,OAS1,OAS2,IFI6,IFIT1,LY96,MX2,HERC6,CREG1,SIGLEC1,EIF2AK2,CEACAM8,C1QB,IFITM1,IFIH1,IFIT2,DDX60,DYSF,ELANE,DEFA4,ABCA1,XAF1,IFITM3,TRIM22,ISG15,LCN2,RNASE6,OLFM4,APOBEC3A,IFI35,RNASE1,VSIG4,MMP9,DHX58,RNASE2,TOP2A,TLR2,LYN,HCK,HBD,MMP8,IRF7,ISG20,ZCCHC2,OLR1,CDC20,GBP2,MS4A4A,DDX58,CCL2,RRM2,SPATS2L,LHFPL2,GBP1,CTSG,CEACAM1,STAT1,CD38,ENTPD1,UBE2J1,TCN2,CXCL10,RTP4,FCER1G,TYMS,B4GALT5,CCNA2,RP2,C3AR1,SAMD9,YIPF6,CARHSP1,TAP1,IFI30,LGALS3BP,TRIM21,FAR2,STIL,PARP12,MFGE8,RNASE3,H2AFZ,CEACAM6,LAP3,YWHAH,GMPR,RIN2,SCARB2,LMNB1,MPO,LCP1,BPGM,TLR4,PRC1,LY6E,CFD,MS4A6A,TPX2,XK,LRRC32,SH3GLB1,TGM2, CTSS,OASL,COL1A2,GINS2,ANP32B,ADAR,RAPGEF5,LYZ,MARCKS,HMGB2,ATP6V1D,SP100,FAM46C,CAMP,COL4A1,TMEM140,ANXA3,FCN1,NETO2,MYOF,EVI2A,FUT4,DNAJC13,CD36,IFI16,POU2AF1,SAMSN1,TCN1,MR1,MNDA,GIMAP4,PRCP,NFE2,CA1,STAT2,PPP1R3D,TACC3,SLPI,C1QA,S100A8,NMI,TLR1,NUSAP1,HLX,ARG1,TMOD1,REXO2,PLA2G4A,PTEN,TNFSF10,NPL,PLEK,GINS3,NOD2,LXN,DNAJC15,HIST1H2AI,MFSD1,DEGS1,FAM46A,CENPN,ITGB2,SCO2,PSMC2,COL15A1,CDC123,DHRS9,HK3,SERPINB1,NOL7,ITGAM 下调基因 CLIC5,ASB1,PTGDS,EPHX2,CDKN1C,TNNC2,KIR2DL3,FCER1A,TGFBR3,FOSB 表 3 交集基因GO富集分析
Table 3. GO enrichment analysis of intersection genes
ONTOLOGY ID 描述 P BP GO:0051607 defense response to virus 0.000 GO:0060337 type I interferon signaling pathway 0.000 GO:0042742 defense response to bacterium 0.000 GO:0019221 cytokine-mediated signaling pathway 0.000 GO:0002221 pattern recognition receptor signaling pathway 0.000 GO:0039529 RIG-I signaling pathway 0.000 GO:0002446 neutrophil mediated immunity 0.000 GO:0002709 toll-like receptor signaling pathway 0.000 GO:0002709 regulation of T cell mediated immunity 0.002 CC GO:0042581 specific granule 0.000 GO:0034774 secretory granule lumen 0.000 GO:0060205 cytoplasmic vesicle lumen 0.000 GO:0005766 primary lysosome 0.000 GO:0045335 phagocytic vesicle 0.001 MF GO:0003725 double-stranded RNA binding 0.000 GO:0008301 DNA binding 0.006 GO:0004518 nuclease activity 0.008 GO:0042379 chemokine receptor binding 0.009 GO:0019207 kinase regulator activity 0.015 表 4 交集基因KEGG信号通路分析
Table 4. KEGG pathway analysis of intersection genes
ID 描述 P hsa05164 Influenza A 0.000 hsa05171 COVID-19 0.000 hsa04621 NOD-like receptor signaling pathway 0.000 hsa04622 RIG-I-like receptor signaling pathway 0.000 hsa05150 Staphylococcus aureus infection 0.001 hsa05322 Systemic lupus erythematosus 0.002 hsa04657 IL-17 signaling pathway 0.004 hsa04623 Cytosolic DNA-sensing pathway 0.016 hsa04610 Complement signaling pathway 0.022 hsa04217 Necroptosis 0.025 hsa04620 Toll-like receptor signaling pathway 0.037 hsa04613 Neutrophil extracellular trap formation 0.044 -
[1] Bastidas Goyes A R,Mora C,Arsanios D M,et al. Systemic lupus erythematosus,a disease conditioned by the environment[J]. Revista Colombiana de Reumatolog í a,2021,28(6):12-20. [2] Barber M R W,Drenkard C,Falasinnu T,et al. Global epidemiology of systemic lupus erythematosus[J]. Nat Rev Rheumatol,2021,17(9):515-532. [3] Gasparotto M,Gatto M,Binda V,et al. Lupus nephritis: Clinical presentations and outcomes in the 21st centu ry[J]. Rheumatology (Oxford),2020,59(Suppl5):v39-v51. [4] Heo YA. Voclosporin: First approval[J]. Drugs,2021,81(5):605-610. [5] Mejia-Vilet J M,Malvar A,Arazi A,et al. The lupus nephritis management renaissance[J]. Kidney Int,2022,101(2):242-255. doi: 10.1016/j.kint.2021.09.012 [6] Parikh S V,Almaani S,Brodsky S,et al. Update on lupus nephritis: Core curriculum 2020[J]. American Journal of Kidney Diseases,2020,76(2):265-281. doi: 10.1053/j.ajkd.2019.10.017 [7] Bernard L,Wang A R,Menez S,et al. Kidney biopsy utility: Patient and clinician perspectives from the kidney precision medicine project[J]. Kidney Med,2023,5(10):100707-100715. doi: 10.1016/j.xkme.2023.100707 [8] Satam H,Joshi K,Mangrolia U,et al. Next-generation sequencing technology: Current trends and advancements[J]. Biology (Basel),2023,12(7):997-1021. doi: 10.3390/biology12070997 [9] Langfelder P,Horvath S. Wgcna: An r package for weighted correlation network analysis[J]. BMC Bioinformatics,2008,9(12):559-571. [10] Nedaie A,Najafi A A. Support vector machine with dirichlet feature mapping[J]. Neural Netw,2017,98(2):87-101. [11] Ubels J,Schaefers T,Punt C,et al. Rainforest: A random forest approach to predict treatment benefit in data from (failed) clinical drug trials[J]. Bioinformatics,2020,36(Suppl_2):i601-i609. [12] Morris A C,Forbes-Osborne M A,Pillai L S,et al. Microarray analysis of xops-mcfp zebrafish retina identifies genes associated with rod photoreceptor degeneration and regeneration[J]. Invest Ophthalmol Vis Sci,2011,52(5):2255-2266. doi: 10.1167/iovs.10-6022 [13] Ko WC C,Li L,Young T R,et al. Gene expression profiling in the skin reveals strong similarities between subacute and chronic cutaneous lupus that are distinct from lupus nephritis[J]. J Invest Dermatol,2021,141(12):2808-2819. doi: 10.1016/j.jid.2021.04.030 [14] Wither J E,Prokopec S D,Noamani B,et al. Identification of a neutrophil-related gene expression signature that is enriched in adult systemic lupus erythematosus patients with active nephritis: Clinical/pathologic associations and etiologic mechanisms[J]. PLoS One,2018,13(5):e0196117. doi: 10.1371/journal.pone.0196117 [15] Berthier C C,Bethunaickan R,Gonzalez-Rivera T,et al. Cross-species transcriptional network analysis defines shared inflammatory responses in murine and human lupus nephritis[J]. J Immunol,2012,189(2):988-1001. [16] Ducreux J,Houssiau F A,Vandepapelière P,et al. Interferon α kinoid induces neutralizing anti-interferon α antibodies that decrease the expression of interferon-induced and b cell activation associated transcripts: Analysis of extended follow-up data from the interferon α kinoid phase i/ii study[J]. Rheumatology (Oxford),2016,55(10):1901-1905. doi: 10.1093/rheumatology/kew262 [17] McKellar J,Moncorgé O,Goujon C. Mx1 proteins and influenza a virus under the microscope[J]. Virologie (Montrouge),2023,27(1):31-34. doi: 10.1684/vir.2023.0990 [18] Tao S,Tan X,Chai W,et al. Knockdown of klf5 ameliorates renal fibrosis in mrl/lpr mice via inhibition of mx1 transcription[J]. Immun Inflamm Dis,2023,11(7):e937. doi: 10.1002/iid3.937 [19] Feng X,Wu H,Grossman J M,et al. Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus eryt hematosus[J]. Arthritis Rheum,2006,54(9):2951-2962. [20] Busse D C,Habgood-Coote D,Clare S,et al. Interferon-induced protein 44 and interferon-induced protein 44-like restrict replication of respiratory syncytial virus[J]. J Virol,2020,94(18):e00297. [21] DeDiego M L,Nogales A,Martinez-Sobrido L,et al. Interferon-induced protein 44 interacts with cellular fk506-binding protein 5,negatively regulates host antiviral responses,and supports virus replication[J]. mBio,2019,10(4):e01839. [22] Philips R L,Wang Y,Cheon H,et al. The jak-stat pathway at 30: Much learned,much more to do[J]. Cell,2022,185(21):3857-3876. doi: 10.1016/j.cell.2022.09.023 [23] Xu W,Chen Z,Shen X,et al. Reno-protective effect of realgar nanoparticles on lupus nephritis of mrl/lpr mice through stat1[J]. Iran J Immunol,2019,16(2):170-181. [24] Fu Y,Xiang Y,Wang Y,et al. The stat1/hmgb1/nf-κb pathway in chronic inflammation and kidney injury after cisplatin exposure[J]. Theranostics,2023,13(9):2757-2773. doi: 10.7150/thno.81406 [25] Kemmner S,Bachmann Q,Steiger S,et al. Stat1 regulates macrophage number and phenotype and prevents renal fibrosis after ischemia-reperfusion injury[J]. American Journal of Physiology-Renal Physiology,2019,316(2):F277-F291. [26] Ding X,Ren Y,He X. Ifn-i mediates lupus nephritis from the beginning to renal fibrosis[J]. Front Immunol,2021,12(4):676082-676099. [27] Arazi A,Rao D A,Berthier C C,et al. The immune cell landscape in kidneys of patients with lupus nephritis[J]. Nature immunology,2019,20(7):902-914. -






 下载:
下载:
 下载:
下载:

