The Role of APOE in Drug Resistance of Colon Cancer Based on Bioinformatics and Cell Experiments
-
摘要:
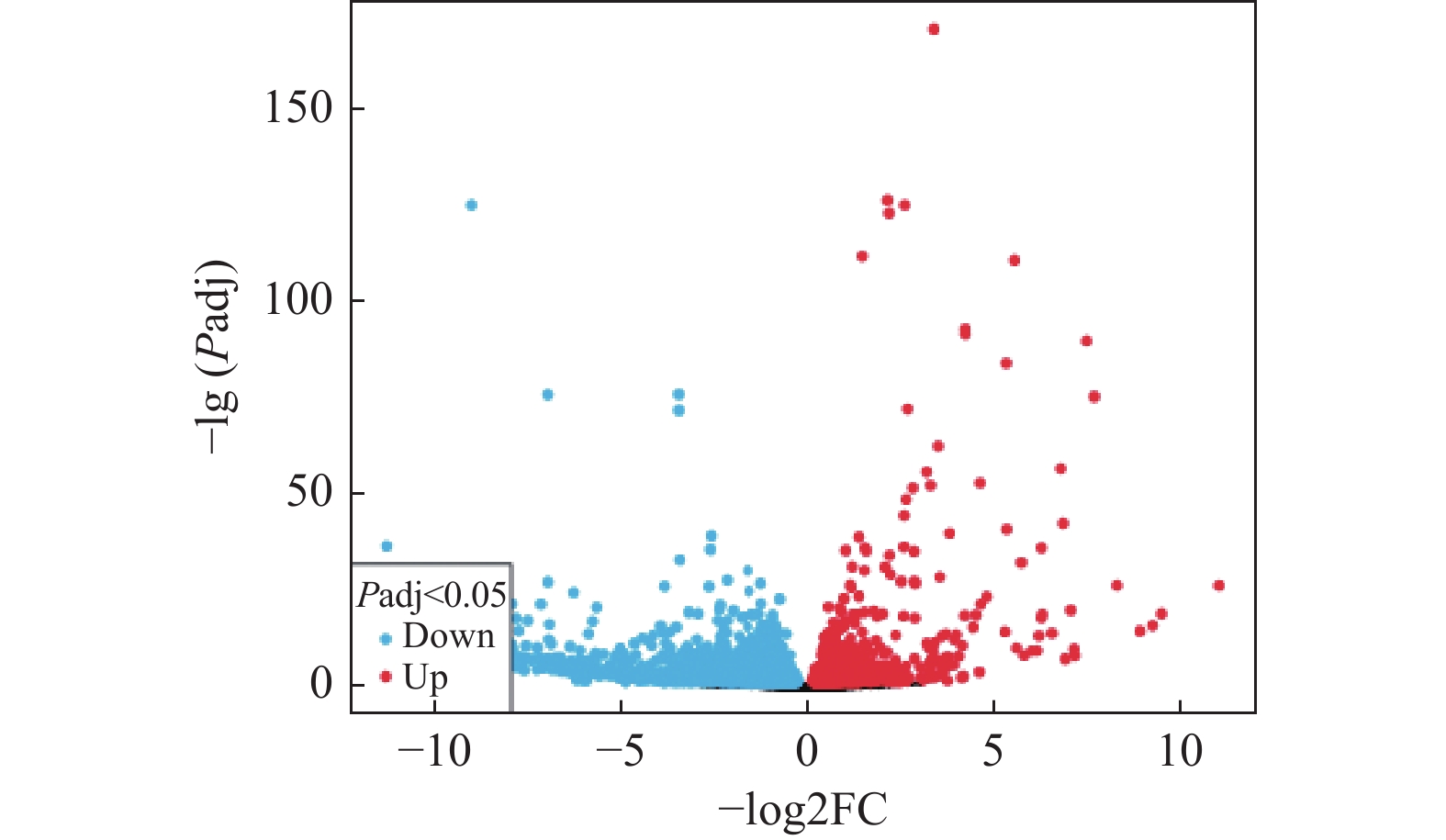
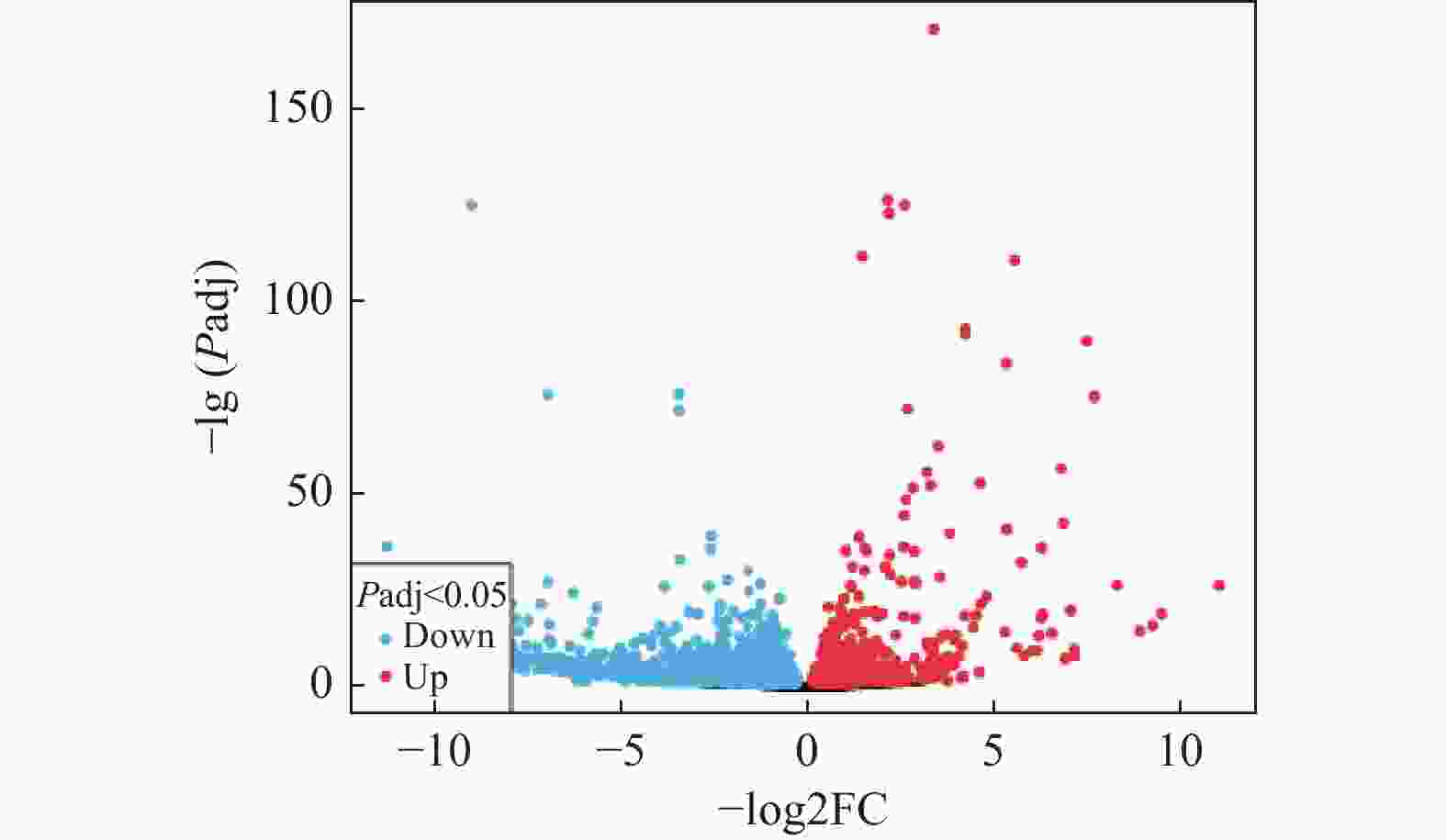
目的 通过生物信息学和细胞实验阐述APOE在结肠癌耐药中的潜在机制。 方法 从GEO数据库下载微阵列数据集GSE196900,利用在线工具GEO2R分析耐药组和对照组之间的差异表达的基因,差异基因进行基因本体论(GO)和京都基因和基因组百科全书(KEGG)途径富集分析。然后构建蛋白-蛋白相互作用(PPI)网络,利用STRING数据库和Cytoscape软件鉴定枢纽基因,以评估枢纽基因在结肠癌中的预后价值。通过Western blod和qRT-pcr检测APOE表达量变化,通过Transwell检测结肠癌细胞的迁移和侵袭能力变化。 结果 GSE196900数据集中得到的差异基因通过GO 分析和KEGG富集分析发现主要集中在受体配体活性和细胞因子−细胞因子受体相互作用通路上。利用Cytoscape软件中的CytoNCA插件进行PPI构建得到10个枢纽的基因,其中,在结肠癌患者中APOE基因的表达量越高患者预后越差(P < 0.05),APOE基因过表达能显著促进结肠癌细胞的迁移和侵袭能力(P < 0.05)。 结论 APOE表达量增加能显著促进结肠癌细胞的迁移和侵袭能力,这可能是APOE基因在结肠癌患者中促进肿瘤进展的机制之一。 Abstract:Objective To evaluate the role and potential mechanism of apolipoprotein E (APOE)in drug resistance of colon cancer by bioinformatic tools and cellular experiments. Methods After downloading the microarray dataset GSE196900 from the GEO database, the online tool GEO2R was used to identify genes that were expressed differently in the drug-resistant and control groups. The differently expressed genes were then examined for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. The STRING database and Cytoscape software were used to build protein-protein interaction (PPI) networks and find hub genes. Hub genes' predictive significance in colon cancer was further assessed. Western blod and qRT-PCR were used to identify changes in APOE expression, whereas Transwell was used to identify changes in the colon cancer cells' capacity for invasion and migration. Results The analysis of GO and KEGG enrichment revealed that the differential genes derived from the GSE196900 dataset were primarily focused on receptor-ligand activity and cytokine-cytokine receptor interaction pathways. Using the CytoNCA plug-in in Cytoscape software, ten hub genes were obtained through PPI construction. Of these, the prognosis of the patients with colon cancer was negatively correlated with the expression of the APOE gene (P < 0.05) and the overexpression of the APOE gene might significantly increase the migration and nvasivenessability of colon cancer cells (P < 0.05). Conclusion The increased expression of APOE significantly promotes the migration and invasion ability of colon cancer cells, which may be one of the mechanisms by which APOE gene promotes tumor progression in the patients with colon cancer. -
Key words:
- Colon cancer /
- Chemotherapy resistance /
- Apolipoprotein E /
- Prognosis /
- GEO
-
图 6 Western blot检测敲低和过表达APOE基因的稳转株表达情况[($ \bar x \pm s $),n = 3]
A:caco-2细胞敲低APOE和RKO细胞过表达APOE蛋白电泳图;B:caco-2细胞敲低APOE和RKO细胞过表达APOE蛋白表达量量化图。NC:空载病毒株,KD:敲低APOE基因稳转细胞株,OE:过表达APOE基因稳转细胞株;***P < 0.001,****P < 0.0001。
Figure 6. The expression of stable transformants with APOE gene knockdown and overexpression was found using Western blot [($ \bar x \pm s $),n = 3]
图 7 qRT-PCR检测敲低和过表达APOE基因的稳转株表达情况[($ \bar x \pm s $),n = 3]
A:caco-2细胞敲低APOE的qRT-PCR实验结果;B:RKO细胞过表达APOE的qRT-PCR实验结果。NC:空载病毒株,KD:敲低APOE基因稳转细胞株,OE:过表达APOE基因稳转细胞株;****P < 0.0001。
Figure 7. The production of stable transformants that overexpressed and knocked down the APOE gene was found using qRT-PCR [($ \bar x \pm s $),n = 3]
图 8 Transwell检测敲低APOE基因之后caco-2细胞的迁移和侵袭能力[($ \bar x \pm s $),n = 3]
A: Transwell实验评估APOE表达量对caco-2细胞迁移和侵袭的影响;B: caco-2细胞Transwell实验数据结果。NC:空载病毒株,KD:敲低APOE基因稳转细胞株;*P < 0.05,**P < 0.01。
Figure 8. Transwell assesses caco-2 cells’ capacity for invasion and migration following APOE gene silencing [($ \bar x \pm s $),n = 3]
图 9 Transwell检测过表达APOE基因之后RKO细胞的迁移和侵袭能力[($ \bar x \pm s $),n = 3]
A:Transwell实验评估APOE表达量对RKO细胞迁移和侵袭的影响;B:RKO细胞Transwell实验数据结果。NC:空载病毒株,OE:过表达APOE基因稳转细胞株;**P < 0.01,***P < 0.001。
Figure 9. Transwell assesses RKO cells' capacity for invasion and migration following overexpression of the APOE gene [($ \bar x \pm s $),n = 3]
表 1 GAPDH和APOE引物序列
Table 1. GAPDH and APOE primer sequences
基因 引物序列(5'-3') 长度 GAPDH F: 5' GGAGCGAGATCCCTCCAAAAT3' 21 R: 5' GGCTGTTGTCATACTTCTCATGG3' 23 APOE F: 5' TTCCTGGCAGGATGCCAGGC3' 20 R: 5' GGTCAGTTGTTCCTCCAGTTC3' 21 -
[1] Rumpold H, Niedersuss-Beke D, Heiler C, et al. Prediction of mortality in metastatic colorectal cancer in a real-life population: A multicenter explorative analysis[J]. BMC Cancer, 2020, 20(1): 1149. [2] Sawicki T, Ruszkowska M, Danielewicz A, et al. A review of colorectal cancer in terms of epidemiology, risk factors, development, symptoms and diagnosis[J]. Cancers (Basel), 2021, 13(9): 2025. [3] Benson A B, Venook A P, Al-Hawary M M, et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology[J]. J Natl Compr Canc Netw, 2021, 19(3): 329-359. [4] Luo X J, Zhao Q, Liu J, et al. Novel genetic and epigenetic biomarkers of prognostic and predictive significance in stage II/III colorectal cancer[J]. Mol Ther, 2021, 29(2): 587-596. [5] Rosenberger G, Li W, Turunen M, et al. Network-based elucidation of colon cancer drug resistance mechanisms by phosphoproteomic time-series analysis[J]. Nat Commun, 2024, 15(1): 3909. [6] Zhang Y, Zheng L. Apolipoprotein: Prospective biomarkers in digestive tract cancer[J]. Transl Cancer Res, 2020, 9(5): 3712-3720. [7] Zhu G X, Gao D, Shao Z Z, et al. Wnt/beta‑catenin signaling: Causes and treatment targets of drug resistance in colorectal cancer (Review)[J]. Mol Med Rep, 2021, 23(2): 105. [8] Peng X, Cai Z, Chen D, et al. Prognostic significance and immune characteristics of APOE in gastric cancer[J]. Aging (Albany NY), 2023, 15(23): 13840-13853. [9] Obradovic A, Chowdhury N, Haake S M, et al. Single-cell protein activity analysis identifies recurrence-associated renal tumor macrophages[J]. Cell, 2021, 184(11): 2988-3005 e16. [10] Lavillegrand J R, Al-Rifai R, Thietart S, et al. Alternating high-fat diet enhances atherosclerosis by neutrophil reprogramming[J]. Nature, 2024, 634(8033): 447-456. [11] Wei Y, Lan B, Zheng T, et al. GSDME-mediated pyroptosis promotes the progression and associated inflammation of atherosclerosis[J]. Nat Commun, 2023, 14(1): 929. [12] Zhao H, Ji Q, Wu Z, et al. Destabilizing heterochromatin by APOE mediates senescence[J]. Nat Aging, 2022, 2(4): 303-316. [13] Zhao N, Ren Y, Yamazaki Y, et al. Alzheimer's risk factors age, APOE genotype, and sex drive distinct molecular pathways[J]. Neuron, 2020, 106(5): 727-42e6. [14] Dai W, Guo C, Wang Y, et al. Identification of hub genes and pathways in lung metastatic colorectal cancer[J]. BMC Cancer, 2023, 23(1): 323. [15] Bonaterra G A, Struck N, Zuegel S, et al. Characterization of atherosclerotic plaques in blood vessels with low oxygenated blood and blood pressure (Pulmonary trunk): role of growth differentiation factor-15 (GDF-15)[J]. BMC Cardiovasc Disord, 2021, 21(1): 601. [16] Huang J, Sun W, Wang Z, et al. FTO suppresses glycolysis and growth of papillary thyroid cancer via decreasing stability of APOE mRNA in an N6-methyladenosine-dependent manner[J]. J Exp Clin Cancer Res, 2022, 41(1): 42. [17] Bancaro N, Cali B, Troiani M, et al. Apolipoprotein E induces pathogenic senescent-like myeloid cells in prostate cancer[J]. Cancer Cell, 2023, 41(3): 602-19e11. [18] Jia Y, Zhang B, Zhang C, et al. Single-Cell transcriptomic analysis of primary and metastatic tumor ecosystems in esophageal squamous cell carcinoma[J]. Adv Sci (Weinh), 2023, 10(7): e2204565. [19] Wan S, He Q Y, Yang Y, et al. SPARC stabilizes APOE to induce cholesterol-dependent invasion and sorafenib resistance in hepatocellular carcinoma[J]. Cancer Res, 2024, 84(11): 1872-1888. [20] Wang Y, Zhu G Q, Yang R, et al. Deciphering intratumoral heterogeneity of hepatocellular carcinoma with microvascular invasion with radiogenomic analysis[J]. J Transl Med, 2023, 21(1): 734. [21] Li J, Gao A, Zhang F, et al. ILT3 promotes tumor cell motility and angiogenesis in non-small cell lung cancer[J]. Cancer Lett, 2021, 501: 263-276. [22] Liu C, Xie J, Lin B, et al. Pan-Cancer single-cell and spatial-resolved profiling reveals the immunosuppressive role of APOE+ macrophages in immune checkpoint inhibitor therapy[J]. Adv Sci (Weinh), 2024, 11(23): e2401061. [23] Chen P, Wang Y, Li J, et al. Diversity and intratumoral heterogeneity in human gallbladder cancer progression revealed by single-cell RNA sequencing[J]. Clin Transl Med, 2021, 11(6): e462. [24] Liu H, Gao J, Feng M, et al. Integrative molecular and spatial analysis reveals evolutionary dynamics and tumor-immune interplay of in situ and invasive acral melanoma[J]. Cancer Cell, 2024, 42(6): 1067-1085. e11. [25] Wong H Y, Sheng Q, Hesterberg A B, et al. Single cell analysis of cribriform prostate cancer reveals cell intrinsic and tumor microenvironmental pathways of aggressive disease[J]. Nat Commun, 2022, 13(1): 6036. -





 下载:
下载: